Exploiting viral vectors to deliver genome editing reagents in plants
- PMID: 38974861
- PMCID: PMC11224180
- DOI: 10.1007/s42994-024-00147-7
Exploiting viral vectors to deliver genome editing reagents in plants
Abstract
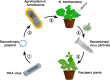
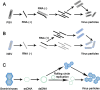
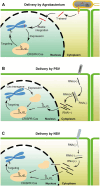
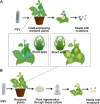
Genome editing holds great promise for the molecular breeding of plants, yet its application is hindered by the shortage of simple and effective means of delivering genome editing reagents into plants. Conventional plant transformation-based methods for delivery of genome editing reagents into plants often involve prolonged tissue culture, a labor-intensive and technically challenging process for many elite crop cultivars. In this review, we describe various virus-based methods that have been employed to deliver genome editing reagents, including components of the CRISPR/Cas machinery and donor DNA for precision editing in plants. We update the progress in these methods with recent successful examples of genome editing achieved through virus-based delivery in different plant species, highlight the advantages and limitations of these delivery approaches, and discuss the remaining challenges.
Keywords: CRISPR/Cas; Genome editing; Plant genome engineering; Virus-based delivery.
© The Author(s) 2024.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures
Similar articles
-
Tools and targets: The dual role of plant viruses in CRISPR-Cas genome editing.Plant Genome. 2023 Jun;16(2):e20220. doi: 10.1002/tpg2.20220. Epub 2022 Jun 14. Plant Genome. 2023. PMID: 35698891 Review.
-
CRISPR/Cas genome editing in plants: Dawn of Agrobacterium transformation for recalcitrant and transgene-free plants for future crop breeding.Plant Physiol Biochem. 2023 Mar;196:724-730. doi: 10.1016/j.plaphy.2023.02.030. Epub 2023 Feb 18. Plant Physiol Biochem. 2023. PMID: 36812799
-
Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes.Mol Plant. 2023 Mar 6;16(3):616-631. doi: 10.1016/j.molp.2023.02.003. Epub 2023 Feb 7. Mol Plant. 2023. PMID: 36751129
-
Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector.Plant J. 2021 Apr;106(2):555-565. doi: 10.1111/tpj.15164. Epub 2021 Mar 10. Plant J. 2021. PMID: 33484202 Free PMC article.
-
Advances in Delivery Mechanisms of CRISPR Gene-Editing Reagents in Plants.Front Genome Ed. 2022 Jan 24;4:830178. doi: 10.3389/fgeed.2022.830178. eCollection 2022. Front Genome Ed. 2022. PMID: 35141701 Free PMC article. Review.
Cited by
-
Genome editing for sustainable agriculture in Peru: advances, potential applications and regulation.Front Genome Ed. 2025 Jun 30;7:1611040. doi: 10.3389/fgeed.2025.1611040. eCollection 2025. Front Genome Ed. 2025. PMID: 40661867 Free PMC article. Review.
-
Engineering an optimized hypercompact CRISPR/Cas12j-8 system for efficient genome editing in plants.Plant Biotechnol J. 2025 Apr;23(4):1153-1164. doi: 10.1111/pbi.14574. Epub 2025 Jan 12. Plant Biotechnol J. 2025. PMID: 39799585 Free PMC article.
-
CRISPR/Cas system-mediated transgene-free or DNA-free genome editing in plants.Theor Appl Genet. 2025 Aug 12;138(9):210. doi: 10.1007/s00122-025-04990-0. Theor Appl Genet. 2025. PMID: 40794116 Review.
-
Protocol for transformation-free genome editing in plants using RNA virus vectors for CRISPR-Cas delivery.STAR Protoc. 2024 Dec 20;5(4):103437. doi: 10.1016/j.xpro.2024.103437. Epub 2024 Nov 5. STAR Protoc. 2024. PMID: 39504248 Free PMC article.
-
Rapid and efficient in planta genome editing in sorghum using foxtail mosaic virus-mediated sgRNA delivery.Plant J. 2025 Jan;121(2):e17196. doi: 10.1111/tpj.17196. Epub 2024 Dec 11. Plant J. 2025. PMID: 39661735 Free PMC article.
References
-
- Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes Nicholas J, Voytas Daniel F, Dinesh-Kumar S, Mahfouz Magdy M. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8:1288–1291. doi: 10.1016/j.molp.2015.02.011. - DOI - PubMed
-
- Altae-Tran H, Kannan S, Demircioglu FE, Oshiro R, Nety SP, McKay LJ, Dlakić M, Inskeep WP, Makarova KS, Macrae RK, Koonin EV, Zhang F. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374:57–65. doi: 10.1126/science.abj6856. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
