A Classical Epithelial State Drives Acute Resistance to KRAS Inhibition in Pancreatic Cancer
- PMID: 38975873
- PMCID: PMC11624508
- DOI: 10.1158/2159-8290.CD-24-0740
A Classical Epithelial State Drives Acute Resistance to KRAS Inhibition in Pancreatic Cancer
Abstract
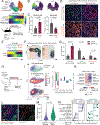
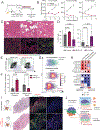
Intratumoral heterogeneity in pancreatic ductal adenocarcinoma (PDAC) is characterized by a balance between basal and classical epithelial cancer cell states, with basal dominance associating with chemoresistance and a dismal prognosis. Targeting oncogenic KRAS, the primary driver of pancreatic cancer, shows early promise in clinical trials, but efficacy is limited by acquired resistance. Using genetically engineered mouse models and patient-derived xenografts, we find that basal PDAC cells are highly sensitive to KRAS inhibitors. Employing fluorescent and bioluminescent reporter systems, we longitudinally track cell-state dynamics in vivo and reveal a rapid, KRAS inhibitor-induced enrichment of the classical state. Lineage tracing uncovers that these enriched classical PDAC cells are a reservoir for disease relapse. Genetic or chemotherapy-mediated ablation of the classical cell state is synergistic with KRAS inhibition, providing a preclinical proof of concept for this therapeutic strategy. Our findings motivate combining classical state-directed therapies with KRAS inhibitors to deepen responses and counteract resistance in pancreatic cancer. Significance: KRAS inhibitors hold promise in pancreatic cancer, but responses are limited by acquired resistance. We find that a classical epithelial cancer cell state is acutely resistant to KRAS inhibition and serves as a reservoir for disease relapse. Targeting the classical state alongside KRAS inhibition deepens responses, revealing a potent therapeutic strategy. See related commentary by Marasco and Misale, p. 2018.
©2024 American Association for Cancer Research.
Figures

References
-
- Arbour KC, Punekar S, Garrido-Laguna I, Hong DS, Wolpin B, Pelster MS, et al. 652O Preliminary clinical activity of RMC-6236, a first-in-class, RAS-selective, tri-complex RAS-MULTI(ON) inhibitor in patients with KRAS mutant pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung cancer (NSCLC). Annals of Oncology 2023;34:S458 doi 10.1016/j.annonc.2023.09.1838. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

