Genome-Wide Gene Birth-Death Dynamics Are Associated with Diet Breadth Variation in Lepidoptera
- PMID: 38976568
- PMCID: PMC11229701
- DOI: 10.1093/gbe/evae095
Genome-Wide Gene Birth-Death Dynamics Are Associated with Diet Breadth Variation in Lepidoptera
Abstract
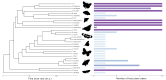
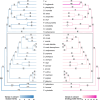
Comparative analyses of gene birth-death dynamics have the potential to reveal gene families that played an important role in the evolution of morphological, behavioral, or physiological variation. Here, we used whole genomes of 30 species of butterflies and moths to identify gene birth-death dynamics among the Lepidoptera that are associated with specialist or generalist feeding strategies. Our work advances this field using a uniform set of annotated proteins for all genomes, investigating associations while correcting for phylogeny, and assessing all gene families rather than a priori subsets. We discovered that the sizes of several important gene families (e.g. those associated with pesticide resistance, xenobiotic detoxification, and/or protein digestion) are significantly correlated with diet breadth. We also found 22 gene families showing significant shifts in gene birth-death dynamics at the butterfly (Papilionoidea) crown node, the most notable of which was a family of pheromone receptors that underwent a contraction potentially linked with a shift to visual-based mate recognition. Our findings highlight the importance of uniform annotations, phylogenetic corrections, and unbiased gene family analyses in generating a list of candidate genes that warrant further exploration.
Keywords: Lepidoptera; butterflies; coevolution; comparative genomics; diet breadth; gene birth–death dynamics; insect–host plant interactions; specialization.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

