Clustered protocadherin cis-interactions are required for combinatorial cell-cell recognition underlying neuronal self-avoidance
- PMID: 38976736
- PMCID: PMC11260096
- DOI: 10.1073/pnas.2319829121
Clustered protocadherin cis-interactions are required for combinatorial cell-cell recognition underlying neuronal self-avoidance
Abstract
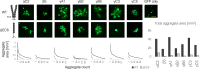
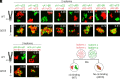
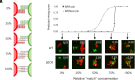
In the developing human brain, only 53 stochastically expressed clustered protocadherin (cPcdh) isoforms enable neurites from individual neurons to recognize and self-avoid while simultaneously maintaining contact with neurites from other neurons. Cell assays have demonstrated that self-recognition occurs only when all cPcdh isoforms perfectly match across the cell boundary, with a single mismatch in the cPcdh expression profile interfering with recognition. It remains unclear, however, how a single mismatched isoform between neighboring cells is sufficient to block erroneous recognitions. Using systematic cell aggregation experiments, we show that abolishing cPcdh interactions on the same membrane (cis) results in a complete loss of specific combinatorial binding between cells (trans). Our computer simulations demonstrate that the organization of cPcdh in linear array oligomers, composed of cis and trans interactions, enhances self-recognition by increasing the concentration and stability of cPcdh trans complexes between the homotypic membranes. Importantly, we show that the presence of mismatched isoforms between cells drastically diminishes the concentration and stability of the trans complexes. Overall, we provide an explanation for the role of the cPcdh assembly arrangements in neuronal self/non-self-discrimination underlying neuronal self-avoidance.
Keywords: adhesion; cadherins; cell–cell recognition specificity; clustered protocadherins; neuronal self-avoidance.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Price S. R., De Marco Garcia N. V., Ranscht B., Jessell T. M., Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell 109, 205–216 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

