SP1 undergoes phase separation and activates RGS20 expression through super-enhancers to promote lung adenocarcinoma progression
- PMID: 38976739
- PMCID: PMC11260144
- DOI: 10.1073/pnas.2401834121
SP1 undergoes phase separation and activates RGS20 expression through super-enhancers to promote lung adenocarcinoma progression
Erratum in
-
Correction for Shan et al., SP1 undergoes phase separation and activates RGS20 expression through super-enhancers to promote lung adenocarcinoma progression.Proc Natl Acad Sci U S A. 2024 Aug 13;121(33):e2414313121. doi: 10.1073/pnas.2414313121. Epub 2024 Aug 5. Proc Natl Acad Sci U S A. 2024. PMID: 39102557 Free PMC article. No abstract available.
Abstract
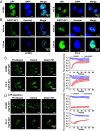
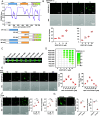
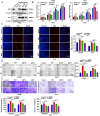
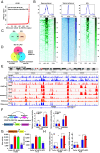
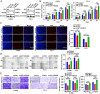
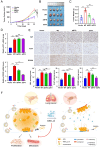
Lung adenocarcinoma (LUAD) is the leading cause of cancer-related death worldwide, but the underlying molecular mechanisms remain largely unclear. The transcription factor (TF) specificity protein 1 (SP1) plays a crucial role in the development of various cancers, including LUAD. Recent studies have indicated that master TFs may form phase-separated macromolecular condensates to promote super-enhancer (SE) assembly and oncogene expression. In this study, we demonstrated that SP1 undergoes phase separation and that its zinc finger 3 in the DNA-binding domain is essential for this process. Through Cleavage Under Targets & Release Using Nuclease (CUT&RUN) using antibodies against SP1 and H3K27ac, we found a significant correlation between SP1 enrichment and SE elements, identified the regulator of the G protein signaling 20 (RGS20) gene as the most likely target regulated by SP1 through SE mechanisms, and verified this finding using different approaches. The oncogenic activity of SP1 relies on its phase separation ability and RGS20 gene activation, which can be abolished by glycogen synthase kinase J4 (GSK-J4), a demethylase inhibitor. Together, our findings provide evidence that SP1 regulates its target oncogene expression through phase separation and SE mechanisms, thereby promoting LUAD cell progression. This study also revealed an innovative target for LUAD therapies through intervening in SP1-mediated SE formation.
Keywords: RGS20; SP1; liquid–liquid phase separation; lung cancer; super-enhancer.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
-
- Siegel R. L., Miller K. D., Wagle N. S., Jemal A., Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023). - PubMed
-
- Bai Q., Wang J., Zhou X., EGFR exon20 insertion mutations in non-small cell lung cancer: Clinical implications and recent advances in targeted therapies. Cancer Treat Rev. 120, 102605 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

