Exploring the potential of Lactocaseibacillus rhamnosus PMC203 in inducing autophagy to reduce the burden of Mycobacterium tuberculosis
- PMID: 38977511
- PMCID: PMC11231020
- DOI: 10.1007/s00430-024-00794-z
Exploring the potential of Lactocaseibacillus rhamnosus PMC203 in inducing autophagy to reduce the burden of Mycobacterium tuberculosis
Abstract
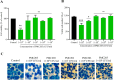
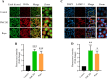
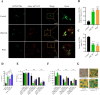
Mycobacterium tuberculosis, a lethal pathogen in human history, causes millions of deaths annually, which demands the development of new concepts of drugs. Considering this fact, earlier research has explored the anti-tuberculosis potential of a probiotic strain, Lactocaseibacillus rhamnosus PMC203, leading to a subsequent focus on the molecular mechanism involved in its effect, particularly on autophagy. In this current study, immunoblotting-based assay exhibited a remarkable expression of autophagy marker LC3-II in the PMC203 treated group compared to an untreated group. A remarkable degradation of p62 was also noticed within treated cells compared to control. Furthermore, the immunofluorescence-based assay showed significant fold change in fluorescence intensity for alexa-647-LC3 and alexa-488-LC3, whereas p62 was degraded noticeably. Moreover, lysosomal biogenesis generation was elevated significantly in terms of LAMP1 and acidic vesicular organelles. As a result, PMC203-induced autophagy played a vital role in reducing M. tuberculosis burden within the macrophages in treated groups compared to untreated group. A colony -forming unit assay also revealed a significant reduction in M. tuberculosis in the treated cells over time. Additionally, the candidate strain significantly upregulated the expression of autophagy induction and lysosomal biogenesis genes. Together, these results could enrich our current knowledge of probiotics-mediated autophagy in tuberculosis and suggest its implications for innovatively managing tuberculosis.
Keywords: Lactocaseibacillus rhamnosus PMC203; Mycobacterium tuberculosis; Anti-tuberculosis; Autophagy; Probiotics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO, GLOBAL TUBERCULOSIS REPORT 2022. Report No. 978–92–4–006172–9, 68 (2022).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

