The caspase-activated DNase promotes cellular senescence
- PMID: 38977850
- PMCID: PMC11329656
- DOI: 10.1038/s44318-024-00163-9
The caspase-activated DNase promotes cellular senescence
Abstract
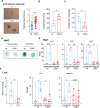
Cellular senescence is a response to many stressful insults. DNA damage is a consistent feature of senescent cells, but in many cases its source remains unknown. Here, we identify the cellular endonuclease caspase-activated DNase (CAD) as a critical factor in the initiation of senescence. During apoptosis, CAD is activated by caspases and cleaves the genomic DNA of the dying cell. The CAD DNase is also activated by sub-lethal signals in the apoptotic pathway, causing DNA damage in the absence of cell death. We show that sub-lethal signals in the mitochondrial apoptotic pathway induce CAD-dependent senescence. Inducers of cellular senescence, such as oncogenic RAS, type-I interferon, and doxorubicin treatment, also depend on CAD presence for senescence induction. By directly activating CAD experimentally, we demonstrate that its activity is sufficient to induce senescence in human cells. We further investigate the contribution of CAD to senescence in vivo and find substantially reduced signs of senescence in organs of ageing CAD-deficient mice. Our results show that CAD-induced DNA damage in response to various stimuli is an essential contributor to cellular senescence.
Keywords: Ageing; Apoptosis; Caspase-activated DNase; Senescence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

