Epigenetic control of skeletal muscle atrophy
- PMID: 38978023
- PMCID: PMC11229277
- DOI: 10.1186/s11658-024-00618-1
Epigenetic control of skeletal muscle atrophy
Abstract
Skeletal muscular atrophy is a complex disease involving a large number of gene expression regulatory networks and various biological processes. Despite extensive research on this topic, its underlying mechanisms remain elusive, and effective therapeutic approaches are yet to be established. Recent studies have shown that epigenetics play an important role in regulating skeletal muscle atrophy, influencing the expression of numerous genes associated with this condition through the addition or removal of certain chemical modifications at the molecular level. This review article comprehensively summarizes the different types of modifications to DNA, histones, RNA, and their known regulators. We also discuss how epigenetic modifications change during the process of skeletal muscle atrophy, the molecular mechanisms by which epigenetic regulatory proteins control skeletal muscle atrophy, and assess their translational potential. The role of epigenetics on muscle stem cells is also highlighted. In addition, we propose that alternative splicing interacts with epigenetic mechanisms to regulate skeletal muscle mass, offering a novel perspective that enhances our understanding of epigenetic inheritance's role and the regulatory network governing skeletal muscle atrophy. Collectively, advancements in the understanding of epigenetic mechanisms provide invaluable insights into the study of skeletal muscle atrophy. Moreover, this knowledge paves the way for identifying new avenues for the development of more effective therapeutic strategies and pharmaceutical interventions.
Keywords: Histone modifications; Skeletal muscle atrophy; Ubiquitin–proteasome; epigenetic; m6A.
© 2024. The Author(s).
Conflict of interest statement
The author declares that no competing interests.
Figures
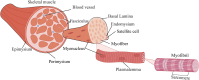
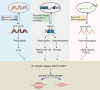

References
-
- Davegardh C, Sall J, Benrick A, Broholm C, Volkov P, Perfilyev A, Henriksen TI, Wu Y, Hjort L, Brons C, et al. VPS39-deficiency observed in type 2 diabetes impairs muscle stem cell differentiation via altered autophagy and epigenetics. Nat Commun. 2021;12(1):2431. doi: 10.1038/s41467-021-22068-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

