YTHDC1 aggravates high glucose-induced retinal vascular endothelial cell injury via m6A modification of CDK6
- PMID: 38978074
- PMCID: PMC11229198
- DOI: 10.1186/s13062-024-00498-7
YTHDC1 aggravates high glucose-induced retinal vascular endothelial cell injury via m6A modification of CDK6
Abstract
Objective: Retinal vascular endothelial cell (RVECs) injury is a major cause of morbidity and mortality among the patients with diabetes. RVECs dysfunction is the predominant pathological manifestation of vascular complication in diabetic retinopathy. N6-methyladenosine (m6A) serves as the most prevalent modification in eukaryotic mRNAs. However, the role of m6A RNA modification in RVECs dysfunction is still unclear.
Methods: RT-qPCR analysis and western blot were conducted to detect the change of m6A RNA modification in diabetic retinopathy. CCK-8 assay, transwell experiment, wound healing assay, tube formation experiment, m6A-IP-qPCR were performed to determine the role of YTHDC1 in RVECs. Retinal trypsin digestion test and H&E staining were used to evaluate histopathological changes.
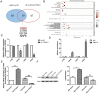
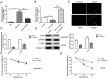
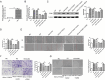
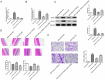
Results: The levels of m6A RNA methylation were significantly up-regulated in HG-induced RVECs, which were caused by increased expression of YTHDC1. YTHDC1 regulated the viability, proliferation, migration and tube formation ability in vitro. YTHDC1 overexpression impaired RVECs function by repressing CDK6 expression, which was mediated by YTHDC1-dependent mRNA decay. Moreover, it showed sh-YTHDC1 inhibited CDK6 nuclear export. Sh-YTHDC1 promotes the mRNA degradation of CDK6 in the nucleus but does not affect the cytoplasmic CDK6 mRNA. In vivo experiments showed that overexpression of CDK6 reversed the protective effect of sh-YTHDC1 on STZ-induced retinal tissue damage.
Conclusion: YTHDC1-mediated m6A methylation regulates diabetes-induced RVECs dysfunction. YTHDC1-CDK6 signaling axis could be therapeutically targeted for treating DR.
Keywords: CDK6; Retinal vascular endothelial cell; YTHDC1; m6A.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- K O et al. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract, 2017. 128(0). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

