Ubiquitin recruiting chimera: more than just a PROTAC
- PMID: 38978100
- PMCID: PMC11232244
- DOI: 10.1186/s13062-024-00497-8
Ubiquitin recruiting chimera: more than just a PROTAC
Abstract
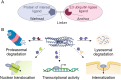
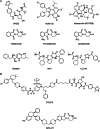
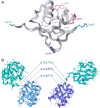
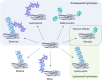
Ubiquitinylation of protein substrates results in various but distinct biological consequences, among which ubiquitin-mediated degradation is most well studied for its therapeutic application. Accordingly, artificially targeted ubiquitin-dependent degradation of various proteins has evolved into the therapeutically relevant PROTAC technology. This tethered ubiquitinylation of various targets coupled with a broad assortment of modifying E3 ubiquitin ligases has been made possible by rational design of bi-specific chimeric molecules that bring these proteins in proximity. However, forced ubiquitinylation inflicted by the binary warheads of a chimeric PROTAC molecule should not necessarily result in protein degradation but can be used to modulate other cellular functions. In this respect it should be noted that the ubiquitinylation of a diverse set of proteins is known to control their transport, transcriptional activity, and protein-protein interactions. This review provides examples of potential PROTAC usage based on non-degradable ubiquitinylation.
Keywords: PROTAC; Protein degradation; Ubiquitin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

