Extracellular vesicle isolation methods identify distinct HIV-1 particles released from chronically infected T-cells
- PMID: 38978287
- PMCID: PMC11231049
- DOI: 10.1002/jev2.12476
Extracellular vesicle isolation methods identify distinct HIV-1 particles released from chronically infected T-cells
Abstract
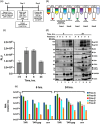
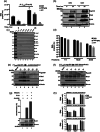
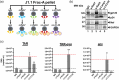
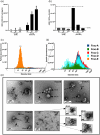
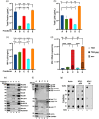
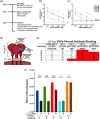
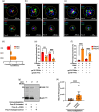
The current study analyzed the intersecting biophysical, biochemical, and functional properties of extracellular particles (EPs) with the human immunodeficiency virus type-1 (HIV-1) beyond the currently accepted size range for HIV-1. We isolated five fractions (Frac-A through Frac-E) from HIV-infected cells by sequential differential ultracentrifugation (DUC). All fractions showed a heterogeneous size distribution with median particle sizes greater than 100 nm for Frac-A through Frac-D but not for Frac-E, which contained small EPs with an average size well below 50 nm. Synchronized and released cultures contained large infectious EPs in Frac-A, with markers of amphisomes and viral components. Additionally, Frac-E uniquely contained EPs positive for CD63, HSP70, and HIV-1 proteins. Despite its small average size, Frac-E contained membrane-protected viral integrase, detectable only after SDS treatment, indicating that it is enclosed in vesicles. Single particle analysis with dSTORM further supported these findings as CD63, HIV-1 integrase, and the viral surface envelope (Env) glycoprotein (gp) colocalized on the same Frac-E particles. Surprisingly, Frac-E EPs were infectious, and infectivity was significantly reduced by immunodepleting Frac-E with anti-CD63, indicating the presence of this protein on the surface of infectious small EPs in Frac-E. To our knowledge, this is the first time that extracellular vesicle (EV) isolation methods have identified infectious small HIV-1 particles (smHIV-1) that are under 50 nm. Collectively, our data indicate that the crossroads between EPs and HIV-1 potentially extend beyond the currently accepted biophysical properties of HIV-1, which may have further implications for viral pathogenesis.
Keywords: HIV‐1; NRTIs; amphisomes; cART; exomeres; exosomes; extracellular particles; extracellular vesicles; smHIV‐1; small HIV.
© 2024 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Anand, S. , Samuel, M. , & Mathivanan, S. (2021). Exomeres: A new member of extracellular vesicles family. Sub‐cellular biochemistry, 97, 89–97. - PubMed
-
- Berman, J. J. (2019). Changing how we think about infectious diseases. Taxonomic Guide to Infectious Diseases, 321–365. 10.1016/B978-0-12-817576-7.00008-0. PMCID: PMC7149514. - DOI
MeSH terms
Substances
Grants and funding
- R21 AI074410/AI/NIAID NIH HHS/United States
- MH134389/NH/NIH HHS/United States
- R21 AI078859/AI/NIAID NIH HHS/United States
- AI127351-01/NH/NIH HHS/United States
- R01 AI043894/AI/NIAID NIH HHS/United States
- AI078859/NH/NIH HHS/United States
- NS099029/NH/NIH HHS/United States
- AI074410/NH/NIH HHS/United States
- W81XWH-11-2-0174/Henry M. Jackson Foundation
- R01 MH134389/MH/NIMH NIH HHS/United States
- AI043894/NH/NIH HHS/United States
- R21 AI127351/AI/NIAID NIH HHS/United States
- R01 NS099029/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

