Genomic epidemiology of Mycobacterium abscessus at an adult cystic fibrosis programme reveals low potential for healthcare-associated transmission
- PMID: 38978544
- PMCID: PMC11228611
- DOI: 10.1183/23120541.00165-2024
Genomic epidemiology of Mycobacterium abscessus at an adult cystic fibrosis programme reveals low potential for healthcare-associated transmission
Abstract
Rationale: Nontuberculous mycobacteria (NTM) has been reported to be transmitted between people with cystic fibrosis (CF) attending CF centres. A suspected Mycobacterium abscessus outbreak was investigated at the University of Texas Southwestern (UTSW) Adult CF Program using a combination of pathogen genomic sequencing and epidemiologic methods. The objectives of the present study were to apply the Healthcare-Associated Links in Transmission of NTM (HALT NTM) study to investigate the occurrence of potential healthcare-associated transmission and/or acquisition of NTM among people with CF infected with genetically similar NTM isolates.
Methods: Whole-genome sequencing of respiratory M. abscessus isolates from 50 people with CF receiving care at UTSW was performed to identify genetically similar isolates. Epidemiologic investigation, comparison of respiratory and environmental isolates, and home residence watershed mapping were studied.
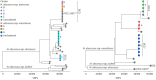
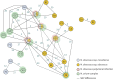
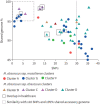
Measurements and main results: Whole-genome sequencing analysis demonstrated seven clusters of genetically similar M. abscessus (four ssp. abscessus and three ssp. massiliense). Epidemiologic investigation revealed potential opportunities for healthcare-associated transmission within three of these clusters. Healthcare environmental sampling did not recover M. abscessus, but did recover four human disease-causing species of NTM. No subjects having clustered infections lived in the same home residence watershed. Some subjects were infected with more than one M. abscessus genotype, both within and outside of the dominant circulating clones.
Conclusions: Healthcare-associated person-to-person transmission of M. abscessus appears to be rare at this centre. However, polyclonal infections of M. abscessus species and subspecies, not originating from the endemic hospital environment, suggest multiple shared modes of acquisition outside the healthcare setting.
Copyright ©The authors 2024.
Conflict of interest statement
Conflict of interest: J.E. Gross reports support for the present manuscript from the Cystic Fibrosis (CF) Foundation and grants or contracts from CF Foundation outside the submitted work; and is a board member of the Rocky Mountain Chapter of the Cystic Fibrosis Foundation and Chair of the American Board of Pediatrics Pulmonology Subboard, disclosures made outside the submitted work. Conflict of interest: J.D. Finklea reports support for the present manuscript from a CF Foundation QI improvement grant, CF Foundation Success with Therapies Research Consortium. Conflict of interest: C.K. Vang reports support for the present manuscript from the CF Foundation. Conflict of interest: J.R. Honda reports support for the present manuscript from the Padosi Foundation and the University of Texas Health Science Center at Tyler; grants or contracts from the National Science Foundation, National Institutes of Health, CF Foundation and the John Chapman Endowed Professorship in Microbiology, outside the submitted work; payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing or educational events from the North American CF Foundation, Yale University, National Jewish Health, NTM Research Consortium and Albert Einstein College of Medicine, outside the submitted work; support for attending meetings and/or travel from the Padosi Foundation, National Science Foundation, National Jewish Health, NIH AGOLD-PRIDE, Albert Einstein College of Medicine, Sam Houston State University, American Thoracic Society, Texas Tech University, Colorado State University, NTM Research Consortium and North American CF Foundation, outside the submitted work; a leadership or fiduciary role for the American Thoracic Society, outside the submitted work; and receipt of equipment, materials, drugs, medical writing, gifts or other services from the University of Texas Health Science Center at Tyler, outside the submitted work. Conflict of interest: M. Strand reports support for the present manuscript from Effort from CF Foundation grant “Prospective Healthcare-Associated Transmission of Nontuberculous Mycobacteria”. Conflict of interest: C.L. Daley reports support for the present manuscript from CF Foundation; grants or contracts from AN2, Bugworks, Insmed, Juvabis and Paratek, outside the submitted work; consulting fees from Genentech and Pfizer, outside the submitted work; and participation on a data safety monitoring or advisory board for AN2, AstraZeneca, Hyfe, Insmed, MannKind, Matinas Biopharma, Nob Hill, Paratek, Spero and Zambon, outside the submitted work. Conflict of interest: The remaining authors have nothing to disclose.
Figures





References
-
- Floto RA, Olivier KN, Saiman L, et al. . US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax 2016; 71: Suppl. 1, i1–i22. doi:10.1136/thoraxjnl-2015-207360 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Medical
