This is a preprint.
A proteomic signature of healthspan
- PMID: 38978645
- PMCID: PMC11230312
- DOI: 10.1101/2024.06.26.24309530
A proteomic signature of healthspan
Update in
-
A proteomic signature of healthspan.Proc Natl Acad Sci U S A. 2025 Jun 10;122(23):e2414086122. doi: 10.1073/pnas.2414086122. Epub 2025 Jun 6. Proc Natl Acad Sci U S A. 2025. PMID: 40478878
Abstract
The focus of aging research has shifted from increasing lifespan to enhancing healthspan to reduce the time spent living with disability. Despite significant efforts to develop biomarkers of aging, few studies have focused on biomarkers of healthspan. We developed a proteomics-based signature of healthspan (healthspan proteomic score (HPS)) using proteomic data from the Olink Explore 3072 assay in the UK Biobank Pharma Proteomics Project (53,018 individuals and 2920 proteins). A lower HPS was associated with higher mortality risk and several age-related conditions, such as COPD, diabetes, heart failure, cancer, myocardial infarction, dementia, and stroke. HPS showed superior predictive accuracy for these outcomes compared to other biological age measures. Proteins associated with HPS were enriched in hallmark pathways such as immune response, inflammation, cellular signaling, and metabolic regulation. The external validity was evaluated using the Essential Hypertension Epigenetics study with proteomic data also from the Olink Explore 3072 and complementary epigenetic data, making it a valuable tool for assessing healthspan and as a potential surrogate marker to complement existing proteomic and epigenetic biological age measures in geroscience-guided studies.
Conflict of interest statement
Conflict of Interest Statement We have no conflicting interests to disclose.
Figures


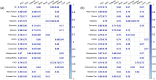
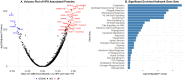
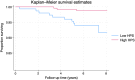
References
-
- GHE: Life expectancy and healthy life expectancy. Available at: https://www.who.int/data/gho/data/themes/mortality-and-global-health-est... [Accessed 21 May 2024].
-
- Khan M., et al. , Global Healthspan Summit 2023: closing the gap between healthspan and lifespan. Nat Aging 4, 445–448 (2024). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
