N6-methyladenosine methylation analysis of circRNAs in acquired middle ear cholesteatoma
- PMID: 38978876
- PMCID: PMC11229040
- DOI: 10.3389/fgene.2024.1396720
N6-methyladenosine methylation analysis of circRNAs in acquired middle ear cholesteatoma
Abstract
Introduction: Middle ear cholesteatoma is a chronic middle ear disease characterized by severe hearing loss and adjacent bone erosion, resulting in numerous complications. This study sought to identify pathways involved in N6-methyladenosine (m6A) modification of circRNA in middle ear cholesteatoma.
Methods: A m6A circRNA epitranscriptomic microarray analysis was performed in middle ear cholesteatoma tissues (n = 5) and normal post-auricular skin samples (n = 5). Bioinformatics analyses subsequently explored the biological functions (Gene Ontology, GO) and signaling pathways (Kyoto Encyclopedia of Genes and Genomes, KEGG) underlying middle ear cholesteatoma pathogenesis. Methylated RNA immunoprecipitation qPCR (MeRIP-qPCR) was performed to verify the presence of circRNAs with m6A modifications in middle ear cholesteatoma and normal skin samples.
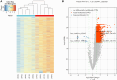
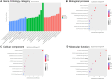
Results: Microarray analysis identified 3,755 circRNAs as significantly differentially modified by m6A methylation in middle ear cholesteatoma compared with the normal post-auricular skin. Among these, 3,742 were hypermethylated (FC ≥ 2, FDR < 0.05) and 13 were hypomethylated (FC ≤ 1/2, FDR < 0.05). GO analysis terms with the highest enrichment score were localization, cytoplasm, and ATP-dependent activity for biological processes, cellular components, and molecular functions respectively. Of the eight hypermethylated circRNA pathways, RNA degradation pathway has the highest enrichment score. Peroxisome Proliferator-Activated Receptor (PPAR) signaling pathway was hypomethylated. To validate the microarray analysis, we conducted MeRIP-qPCR to assess the methylation levels of five specific m6A-modified circRNAs: hsa_circRNA_061554, hsa_circRNA_001454, hsa_circRNA_031526, hsa_circRNA_100833, and hsa_circRNA_022382. The validation was highly consistent with the findings from the microarray analysis.
Conclusion: Our study firstly presents m6A modification patterns of circRNAs in middle ear cholesteatoma. This finding suggests a direction for circRNA m6A modification research in the etiology of cholesteatoma and provides potential therapeutic targets for the treatment of middle ear cholesteatoma.
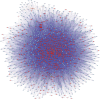
Keywords: circRNA-miRNA-mRNA network; circular RNAs (circRNAs); m6A modification; microarray analysis; middle ear cholesteatoma.
Copyright © 2024 He, Mahmoudi, Yao, Yuan, Fu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

