This is a preprint.
Skp1 is a conserved structural component of the meiotic synaptonemal complex
- PMID: 38979327
- PMCID: PMC11230192
- DOI: 10.1101/2024.06.24.600447
Skp1 is a conserved structural component of the meiotic synaptonemal complex
Abstract
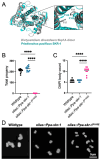
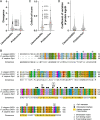
The synaptonemal complex (SC) is a meiotic interface that assembles between parental chromosomes and is essential for the formation of gametes. While the dimensions and ultrastructure of the SC are conserved across eukaryotes, its protein components are highly divergent. Recently, an unexpected component of the SC has been described in the nematode C. elegans: the Skp1-related proteins SKR-1/2, which are components of the Skp1, Cullin, F-box (SCF) ubiquitin ligase. Here, we find that the role of SKR-1 in the SC is conserved in nematodes. The P. pacificus Skp1 ortholog, Ppa-SKR-1, colocalizes with other SC proteins throughout meiotic prophase, where it occupies the middle of the SC. Like in C. elegans, the dimerization interface of Ppa-SKR-1 is required for its SC function. A dimerization mutant, Ppa-skr-1 F105E , fails to assemble SC and is almost completely sterile. Interestingly, the evolutionary trajectory of SKR-1 contrasts with other SC proteins. Unlike most SC proteins, SKR-1 is highly conserved in nematodes. Our results suggest that the structural role of SKR-1 in the SC has been conserved since the common ancestor of C. elegans and P. pacificus, and that rapidly evolving SC proteins have maintained the ability to interact with SKR-1 for at least 100 million years.
Keywords: P. pacificus; SCF; Skp1; evolution; meiosis; nematodes; synaptonemal complex.
Figures


Similar articles
-
The structural role of Skp1 in the synaptonemal complex is conserved in nematodes.Genetics. 2024 Nov 6;228(3):iyae153. doi: 10.1093/genetics/iyae153. Genetics. 2024. PMID: 39293001 Free PMC article.
-
Skp1 proteins are structural components of the synaptonemal complex in C. elegans.Sci Adv. 2024 Feb 16;10(7):eadl4876. doi: 10.1126/sciadv.adl4876. Epub 2024 Feb 14. Sci Adv. 2024. PMID: 38354250 Free PMC article.
-
The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis.Curr Biol. 2002 Feb 19;12(4):277-87. doi: 10.1016/s0960-9822(02)00682-6. Curr Biol. 2002. PMID: 11864567
-
The organization, regulation, and biological functions of the synaptonemal complex.Asian J Androl. 2021 Nov-Dec;23(6):580-589. doi: 10.4103/aja202153. Asian J Androl. 2021. PMID: 34528517 Free PMC article. Review.
-
The synaptonemal complex of basal metazoan hydra: more similarities to vertebrate than invertebrate meiosis model organisms.J Genet Genomics. 2014 Mar 20;41(3):107-15. doi: 10.1016/j.jgg.2014.01.009. Epub 2014 Feb 20. J Genet Genomics. 2014. PMID: 24656231 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
