This is a preprint.
Suppression of astrocyte BMP signaling improves fragile X syndrome molecular signatures and functional deficits
- PMID: 38979341
- PMCID: PMC11230279
- DOI: 10.1101/2024.06.19.599752
Suppression of astrocyte BMP signaling improves fragile X syndrome molecular signatures and functional deficits
Abstract
Fragile X syndrome (FXS) is a monogenic neurodevelopmental disorder with manifestations spanning molecular, neuroanatomical, and behavioral changes. Astrocytes contribute to FXS pathogenesis and show hundreds of dysregulated genes and proteins; targeting upstream pathways mediating astrocyte changes in FXS could therefore be a point of intervention. To address this, we focused on the bone morphogenetic protein (BMP) pathway, which is upregulated in FXS astrocytes. We generated a conditional KO (cKO) of Smad4 in astrocytes to suppress BMP signaling, and found this lessens audiogenic seizure severity in FXS mice. To ask how this occurs on a molecular level, we performed in vivo transcriptomic and proteomic profiling of cortical astrocytes, finding upregulation of metabolic pathways, and downregulation of secretory machinery and secreted proteins in FXS astrocytes, with these alterations no longer present when BMP signaling is suppressed. Functionally, astrocyte Smad4 cKO restores deficits in inhibitory synapses present in FXS auditory cortex. Thus, astrocytes contribute to FXS molecular and functional phenotypes, and targeting astrocytes can mitigate FXS symptoms.
Conflict of interest statement
Competing Interests The authors have no competing interests to declare.
Figures



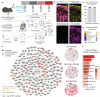


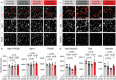
Similar articles
-
Selective Deletion of Astroglial FMRP Dysregulates Glutamate Transporter GLT1 and Contributes to Fragile X Syndrome Phenotypes In Vivo.J Neurosci. 2016 Jul 6;36(27):7079-94. doi: 10.1523/JNEUROSCI.1069-16.2016. J Neurosci. 2016. PMID: 27383586 Free PMC article.
-
Astrocyte-mediated purinergic signaling is upregulated in a mouse model of Fragile X syndrome.Glia. 2021 Jul;69(7):1816-1832. doi: 10.1002/glia.23997. Epub 2021 Mar 22. Glia. 2021. PMID: 33754385
-
Astrocyte-secreted thrombospondin-1 modulates synapse and spine defects in the fragile X mouse model.Mol Brain. 2016 Aug 2;9(1):74. doi: 10.1186/s13041-016-0256-9. Mol Brain. 2016. PMID: 27485117 Free PMC article.
-
Astrocytes in fragile X syndrome.Front Cell Neurosci. 2024 Jan 8;17:1322541. doi: 10.3389/fncel.2023.1322541. eCollection 2023. Front Cell Neurosci. 2024. PMID: 38259499 Free PMC article. Review.
-
Auditory processing in fragile x syndrome.Front Cell Neurosci. 2014 Feb 4;8:19. doi: 10.3389/fncel.2014.00019. eCollection 2014. Front Cell Neurosci. 2014. PMID: 24550778 Free PMC article. Review.
References
-
- Hagerman R. J. et al. Fragile X syndrome. Nat. Rev. Dis. Primer 3, (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
