Anandamide modulation of monocyte-derived Langerhans cells: implications for immune homeostasis and skin inflammation
- PMID: 38979427
- PMCID: PMC11228147
- DOI: 10.3389/fimmu.2024.1423776
Anandamide modulation of monocyte-derived Langerhans cells: implications for immune homeostasis and skin inflammation
Abstract
Introduction: The endocannabinoid system (ECS), named after the chemical compounds found in the cannabis plant, is a regulatory network of neurotransmitters, receptors, and enzymes that plays crucial roles in skin health and disease. Endogenous ligands of the ECS, called endocannabinoids, have proven to be important regulators of immune responses. One of the most prevalent endocannabinoids, arachidonoylethanolamide (also known as anandamide), is known for its anti-inflammatory effects. Langerhans cells (LCs) are the sole antigen-presenting cells present in the human epidermis. They serve as the first line of defense against pathogens and are essential for the skin's specific immune responses and play a critical role in maintaining tissue homeostasis; however, little is known about the effect of endocannabinoids on these cells. Our research aimed to provide the connection between monocyte-derived Langerhans cells (moLCs) and the ECS, shedding light on their collaborative roles in immune homeostasis and inflammation.
Methods: Human monocytes were differentiated into moLCs using established protocols. Anandamide was applied during the differentiation process to test its effect on the viability, marker expression, and cytokine production of the cells, as well as in short term treatments for intracellular calcium measurement. TLR ligands applied after the differentiation protocol were used to activate moLCs. The impact of anandamide on the functionality of moLCs was further assessed using differential gene expression analysis of bulk RNA-Seq data, moLC-T cell cocultures, while ELISpot was employed to determine polarization of T cells activated in the aforementioned cocultures.
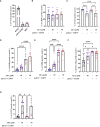
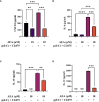
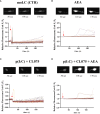
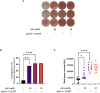
Results: Anandamide did not significantly affect the viability of moLCs up to 10 µM. When applied during the differentiation process it had only a negligible effect on CD207 expression, the prototypic marker of LCs; however, there was an observed reduction in CD1a expression by moLCs. Anandamide had no significant effects on the maturation status of moLCs, nor did it affect the maturation induced by TLR3 and TLR7/8 agonists. MoLCs differentiated in the presence of anandamide did however show decreased production of CXCL8, IL-6, IL-10 and IL-12 cytokines induced by TLR3 and TLR7/8 activation. Anandamide-treated moLCs showed an increased capability to activate naïve T cells; however, not to the level seen with combined TLR agonism. RNA sequencing analysis of moLCs differentiated with anandamide showed modest changes compared to control cells but did reveal an inhibitory effect on oxidative phosphorylation specifically in activated moLCs. Anandamide also promoted the polarization of naïve T cells towards a Th1 phenotype.
Discussion: Our results show that anandamide has nuanced effects on the differentiation, maturation, cytokine secretion, metabolism and function of activated moLCs. Among these changes the decrease in CD1a expression on moLCs holds promise to selectively dampen inflammation induced by CD1a restricted T cells, which have been implicated as drivers of inflammation in common inflammatory skin conditions such as psoriasis, atopic dermatitis and contact dermatitis.
Keywords: CD1a; Langerhans; anandamide; cells; epidermis; skin immunology.
Copyright © 2024 Pénzes, Horváth, Molnár, Fekete, Pázmándi, Bácsi and Szöllősi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Natriuretic peptides modulate monocyte-derived Langerhans cell differentiation and promote a migratory phenotype.Front Immunol. 2025 Jun 9;16:1593141. doi: 10.3389/fimmu.2025.1593141. eCollection 2025. Front Immunol. 2025. PMID: 40552299 Free PMC article.
-
Sphingosine 1-phospate differentially modulates maturation and function of human Langerhans-like cells.J Dermatol Sci. 2016 Apr;82(1):9-17. doi: 10.1016/j.jdermsci.2016.01.002. Epub 2016 Jan 9. J Dermatol Sci. 2016. PMID: 26803226
-
Differentiation of Langerhans Cells from Monocytes and Their Specific Function in Inducing IL-22-Specific Th Cells.J Immunol. 2018 Nov 15;201(10):3006-3016. doi: 10.4049/jimmunol.1701402. Epub 2018 Oct 15. J Immunol. 2018. PMID: 30322965 Free PMC article.
-
Notch-Mediated Generation of Monocyte-Derived Langerhans Cells: Phenotype and Function.J Invest Dermatol. 2021 Jan;141(1):84-94.e6. doi: 10.1016/j.jid.2020.05.098. Epub 2020 Jun 6. J Invest Dermatol. 2021. PMID: 32522485 Free PMC article. Review.
-
Uncovering the Mysteries of Langerhans Cells, Inflammatory Dendritic Epidermal Cells, and Monocyte-Derived Langerhans Cell-Like Cells in the Epidermis.Front Immunol. 2018 Jul 30;9:1768. doi: 10.3389/fimmu.2018.01768. eCollection 2018. Front Immunol. 2018. PMID: 30105033 Free PMC article. Review.
Cited by
-
Natriuretic peptides modulate monocyte-derived Langerhans cell differentiation and promote a migratory phenotype.Front Immunol. 2025 Jun 9;16:1593141. doi: 10.3389/fimmu.2025.1593141. eCollection 2025. Front Immunol. 2025. PMID: 40552299 Free PMC article.
-
Atopic dermatitis: pathogenesis and therapeutic intervention.MedComm (2020). 2024 Dec 8;5(12):e70029. doi: 10.1002/mco2.70029. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39654684 Free PMC article. Review.
-
The endocannabinoid anandamide prevents TH17 programming of activated T lymphocytes while preserving TH1 responses.Front Pharmacol. 2024 Dec 20;15:1528759. doi: 10.3389/fphar.2024.1528759. eCollection 2024. Front Pharmacol. 2024. PMID: 39759451 Free PMC article.
-
Decoding Macrophage Dynamics: A Pathway to Understanding and Treating Inflammatory Skin Diseases.Int J Mol Sci. 2025 May 1;26(9):4287. doi: 10.3390/ijms26094287. Int J Mol Sci. 2025. PMID: 40362523 Free PMC article. Review.
References
-
- Devane WA, Dysarz FA, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol Pharmacol. (1988) 34:605–13. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

