Immunological correlates of protection mediated by a whole organism, Cryptococcus neoformans, vaccine deficient in chitosan
- PMID: 38980038
- PMCID: PMC11323574
- DOI: 10.1128/mbio.01746-24
Immunological correlates of protection mediated by a whole organism, Cryptococcus neoformans, vaccine deficient in chitosan
Abstract
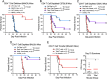
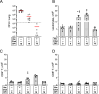
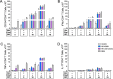
The global burden of infections due to the pathogenic fungus Cryptococcus is substantial in persons with low CD4+ T-cell counts. Previously, we deleted three chitin deacetylase genes from Cryptococcus neoformans to create a chitosan-deficient, avirulent strain, designated as cda1∆2∆3∆, which, when used as a vaccine, protected mice from challenge with virulent C. neoformans strain KN99. Here, we explored the immunological basis for protection. Vaccine-mediated protection was maintained in mice lacking B cells or CD8+ T cells. In contrast, protection was lost in mice lacking α/β T cells or CD4+ T cells. Moreover, CD4+ T cells from vaccinated mice conferred protection upon adoptive transfer to naive mice. Importantly, while monoclonal antibody-mediated depletion of CD4+ T cells just prior to vaccination resulted in complete loss of protection, significant protection was retained in mice depleted of CD4+ T cells after vaccination but prior to challenge. Vaccine-mediated protection was lost in mice genetically deficient in interferon-γ (IFNγ), tumor necrosis factor alpha (TNFα), or interleukin (IL)-23p19. A robust influx of leukocytes and IFNγ- and TNFα-expressing CD4+ T cells was seen in the lungs of vaccinated and challenged mice. Finally, a higher level of IFNγ production by lung cells stimulated ex vivo correlated with lower fungal burden in the lungs. Thus, while B cells and CD8+ T cells are dispensable, IFNγ and CD4+ T cells have overlapping roles in generating protective immunity prior to cda1∆2∆3∆ vaccination. However, once vaccinated, protection becomes less dependent on CD4+ T cells, suggesting a strategy for vaccinating HIV+ persons prior to loss of CD4+ T cells.
Importance: The fungus Cryptococcus neoformans is responsible for >100,000 deaths annually, mostly in persons with impaired CD4+ T-cell function such as AIDS. There are no approved human vaccines. We previously created a genetically engineered avirulent strain of C. neoformans, designated as cda1∆2∆3∆. When used as a vaccine, cda1∆2∆3∆ protects mice against a subsequent challenge with a virulent C. neoformans strain. Here, we defined components of the immune system responsible for vaccine-mediated protection. We found that while B cells and CD8+ T cells were dispensible, protection was lost in mice genetically deficient in CD4+ T cells and the cytokines IFNγ, TNFα, or IL-23. A robust influx of cytokine-producing CD4+ T cells was seen in the lungs of vaccinated mice following infection. Importantly, protection was retained in mice depleted of CD4+ T cells following vaccination, suggesting a strategy to protect persons who are at risk of future CD4+ T-cell dysfunction.
Keywords: AIDS; Cryptococcus neoformans; T cells; live vector vaccines; mycology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
Immunological correlates of protection mediated by a whole organism Cryptococcus neoformans vaccine deficient in chitosan.bioRxiv [Preprint]. 2024 Jun 14:2024.06.12.598760. doi: 10.1101/2024.06.12.598760. bioRxiv. 2024. Update in: mBio. 2024 Aug 14;15(8):e0174624. doi: 10.1128/mbio.01746-24. PMID: 38915489 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
