Polymorphism-Aware Models in RevBayes: Species Trees, Disentangling Balancing Selection, and GC-Biased Gene Conversion
- PMID: 38980178
- PMCID: PMC11272101
- DOI: 10.1093/molbev/msae138
Polymorphism-Aware Models in RevBayes: Species Trees, Disentangling Balancing Selection, and GC-Biased Gene Conversion
Abstract
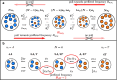
The role of balancing selection is a long-standing evolutionary puzzle. Balancing selection is a crucial evolutionary process that maintains genetic variation (polymorphism) over extended periods of time; however, detecting it poses a significant challenge. Building upon the Polymorphism-aware phylogenetic Models (PoMos) framework rooted in the Moran model, we introduce a PoMoBalance model. This novel approach is designed to disentangle the interplay of mutation, genetic drift, and directional selection (GC-biased gene conversion), along with the previously unexplored balancing selection pressures on ultra-long timescales comparable with species divergence times by analyzing multi-individual genomic and phylogenetic divergence data. Implemented in the open-source RevBayes Bayesian framework, PoMoBalance offers a versatile tool for inferring phylogenetic trees as well as quantifying various selective pressures. The novel aspect of our approach in studying balancing selection lies in polymorphism-aware phylogenetic models' ability to account for ancestral polymorphisms and incorporate parameters that measure frequency-dependent selection, allowing us to determine the strength of the effect and exact frequencies under selection. We implemented validation tests and assessed the model on the data simulated with SLiM and a custom Moran model simulator. Real sequence analysis of Drosophila populations reveals insights into the evolutionary dynamics of regions subject to frequency-dependent balancing selection, particularly in the context of sex-limited color dimorphism in Drosophila erecta.
Keywords: Bayesian inference with MCMC; GC-biased gene conversion; balancing selection; polymorphism-aware phylogenetic models; site frequency spectrum; species trees.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of interest statement None declared.
Figures







Similar articles
-
Linking great apes genome evolution across time scales using polymorphism-aware phylogenetic models.Mol Biol Evol. 2013 Oct;30(10):2249-62. doi: 10.1093/molbev/mst131. Epub 2013 Aug 1. Mol Biol Evol. 2013. PMID: 23906727 Free PMC article.
-
A population genetics-phylogenetics approach to inferring natural selection in coding sequences.PLoS Genet. 2011 Dec;7(12):e1002395. doi: 10.1371/journal.pgen.1002395. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144911 Free PMC article.
-
Interaction between selection and biased gene conversion in mammalian protein-coding sequence evolution revealed by a phylogenetic covariance analysis.Mol Biol Evol. 2013 Feb;30(2):356-68. doi: 10.1093/molbev/mss231. Epub 2012 Sep 29. Mol Biol Evol. 2013. PMID: 23024185
-
Evolution of balanced genetic polymorphism.Mol Ecol. 2000 Dec;9(12):1953-63. doi: 10.1046/j.1365-294x.2000.01125.x. Mol Ecol. 2000. PMID: 11123608 Review.
-
Rapid evolutionary change, constraints and the maintenance of polymorphism in natural populations of Drosophila melanogaster.Mol Ecol. 2024 May;33(10):e17024. doi: 10.1111/mec.17024. Epub 2023 May 24. Mol Ecol. 2024. PMID: 37222070 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

