Exploring COVID-19 Pandemic Disparities with Transcriptomic Meta-analysis from the Perspective of Personalized Medicine
- PMID: 38980578
- PMCID: PMC11436439
- DOI: 10.1007/s12275-024-00154-9
Exploring COVID-19 Pandemic Disparities with Transcriptomic Meta-analysis from the Perspective of Personalized Medicine
Abstract
Infection with SARS-CoV2, which is responsible for COVID-19, can lead to differences in disease development, severity and mortality rates depending on gender, age or the presence of certain diseases. Considering that existing studies ignore these differences, this study aims to uncover potential differences attributable to gender, age and source of sampling as well as viral load using bioinformatics and multi-omics approaches. Differential gene expression analyses were used to analyse the phenotypic differences between SARS-CoV-2 patients and controls at the mRNA level. Pathway enrichment analyses were performed at the gene set level to identify the activated pathways corresponding to the differences in the samples. Drug repurposing analysis was performed at the protein level, focusing on host-mediated drug candidates to uncover potential therapeutic differences. Significant differences (i.e. the number of differentially expressed genes and their characteristics) were observed for COVID-19 at the mRNA level depending on the sample source, gender and age of the samples. The results of the pathway enrichment show that SARS-CoV-2 can be combated more effectively in the respiratory tract than in the blood samples. Taking into account the different sample sources and their characteristics, different drug candidates were identified. Evaluating disease prediction, prevention and/or treatment strategies from a personalised perspective is crucial. In this study, we not only evaluated the differences in COVID-19 from a personalised perspective, but also provided valuable data for further experimental and clinical efforts. Our findings could shed light on potential pandemics.
Keywords: Bioinformatics; COVID-19; Gender and age disparities; Host-directed drug candidates; Personalized medicine; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors report there are no competing interests to declare.
Figures


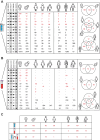


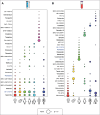
References
-
- Acer, Ö., Genç Bahçe, Y., & Özüdoğru, O. (2022). Association of viral load with age, gender, disease severity, and death in severe acute respiratory syndrome coronavirus 2 variants. Journal of Medical Virology,94, 3063–3069. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

