Microglia Gravitate toward Amyloid Plaques Surrounded by Externalized Phosphatidylserine via TREM2
- PMID: 38981007
- PMCID: PMC11425970
- DOI: 10.1002/advs.202400064
Microglia Gravitate toward Amyloid Plaques Surrounded by Externalized Phosphatidylserine via TREM2
Abstract
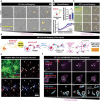
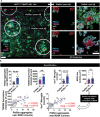
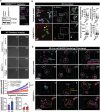
Microglia play a crucial role in synaptic elimination by engulfing dystrophic neurons via triggering receptors expressed on myeloid cells 2 (TREM2). They are also involved in the clearance of beta-amyloid (Aβ) plaques in Alzheimer's disease (AD); nonetheless, the driving force behind TREM2-mediated phagocytosis of beta-amyloid (Aβ) plaques remains unknown. Here, using advanced 2D/3D/4D co-culture systems with loss-of-function mutations in TREM2 (a frameshift mutation engineered in exon 2) brain organoids/microglia/assembloids, it is identified that the clearance of Aβ via TREM2 is accelerated by externalized phosphatidylserine (ePtdSer) generated from dystrophic neurons surrounding the Aβ plaques. Moreover, it is investigated whether microglia from both sporadic (CRISPR-Cas9-based APOE4 lines) and familial (APPNL-G-F/MAPT double knock-in mice) AD models show reduced levels of TREM2 and lack of phagocytic activity toward ePtdSer-positive Aβ plaques. Herein new insight is provided into TREM2-dependent microglial phagocytosis of Aβ plaques in the context of the presence of ePtdSer during AD progression.
Keywords: Alzheimer's disease; TREM2; beta‐amyloid; microglia; phosphatidylserine.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Paolicelli R. C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T. A., Guiducci E., Dumas L., Ragozzino D., Gross C. T., Science 2011, 333, 1456. - PubMed
-
- a) Streit W. J., Glia 2002, 40, 133; - PubMed
- b) Zhou X. G., Qiu W. Q., Yu L., Pan R., Teng J. F., Sang Z. P., Law B. Y., Zhao Y., Zhang L., Yan L., Tang Y., Sun X. L., Wong V. K. W., Yu C. L., Wu J. M., Qin D. L., Wu A. G., Inflamm. Regen. 2022, 42, 25; - PMC - PubMed
- c) Baik S. H., Kang S., Son S. M., Mook‐Jung I., Glia 2016, 64, 2274. - PubMed
-
- a) Kurematsu C., Sawada M., Ohmuraya M., Tanaka M., Kuboyama K., Ogino T., Matsumoto M., Oishi H., Inada H., Ishido Y., Sakakibara Y., Nguyen H. B., Thai T. Q., Kohsaka S., Ohno N., Yamada M. K., Asai M., Sokabe M., Nabekura J., Asano K., Tanaka M., Sawamoto K., J. Exp. Med. 2022, 219, e20202304; - PMC - PubMed
- b) Scott‐Hewitt N., Perrucci F., Morini R., Erreni M., Mahoney M., Witkowska A., Carey A., Faggiani E., Schuetz L. T., Mason S., Tamborini M., Bizzotto M., Passoni L., Filipello F., Jahn R., Stevens B., Matteoli M., EMBO J. 2020, 39, 105380; - PMC - PubMed
- c) Shacham‐Silverberg V., Sar Shalom H., Goldner R., Golan‐Vaishenker Y., Gurwicz N., Gokhman I., Yaron A., Cell Death Dis. 2018, 9, 1116. - PMC - PubMed
-
- a) Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J. S., Younkin S., Hazrati L., Collinge J., Pocock J., Lashley T., Williams J., Lambert J. C., Amouyel P., Goate A., Rademakers R., Morgan K., Powell J., St George‐Hyslop P., Singleton A., Hardy J., Alzheimer Genetic Analysis G., N Engl. J. Med. 2013, 368, 117; - PubMed
- b) Rueda‐Carrasco J., Sokolova D., Lee S‐E., Childs T., Jurčáková N., Crowley G., De Schepper S., Ge J. Z., Lachica J. I., Toomey C. E., Freeman O. J., Hardy J., Barnes S. J., Lashley T., Stevens B., Chang S., Hong S., EMBO J. 2023, 42, 113246. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- NRF-2022R1C1C2012736/National Research Foundation of Korea
- RS-2023-00266110/National Research Foundation of Korea
- NRF002746441G0003021/National Research Foundation of Korea
- NRF-I21SS7606036/National Research Foundation of Korea
- RS-2020-KH106747/Korea Health Industry Development Institute/Republic of Korea
- RS-2020-KH106773/Korea Health Industry Development Institute/Republic of Korea
- RS-2022-KH126808/Korea Health Industry Development Institute/Republic of Korea
- RS-2024-00339665/Korea Health Industry Development Institute/Republic of Korea
- RS-2020-KH106747/Korea Dementia Research Center
- RS-2020-KH106773/Korea Dementia Research Center
- RS-2022-KH126808/Korea Dementia Research Center
- RS-2024-00339665/Korea Dementia Research Center
- RS-2020-KH106747/Ministry of Health & Welfare and Ministry of science and ICT
- RS-2020-KH106773/Ministry of Health & Welfare and Ministry of science and ICT
- RS-2022-KH126808/Ministry of Health & Welfare and Ministry of science and ICT
- RS-2024-00339665/Ministry of Health & Welfare and Ministry of science and ICT
- Republic of Korea
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
