Antibody-assisted selective isolation of Purkinje cell nuclei from mouse cerebellar tissue
- PMID: 38981474
- PMCID: PMC11294835
- DOI: 10.1016/j.crmeth.2024.100816
Antibody-assisted selective isolation of Purkinje cell nuclei from mouse cerebellar tissue
Abstract
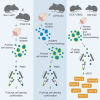
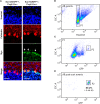
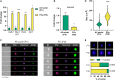
We developed a method that utilizes fluorescent labeling of nuclear envelopes alongside cytometry sorting for the selective isolation of Purkinje cell (PC) nuclei. Beginning with SUN1 reporter mice, we GFP-tagged envelopes to confirm that PC nuclei could be accurately separated from other cell types. We then developed an antibody-based protocol to make PC nuclear isolation more robust and adaptable to cerebellar tissues of any genotypic background. Immunofluorescent labeling of the nuclear membrane protein RanBP2 enabled the isolation of PC nuclei from C57BL/6 cerebellum. By analyzing the expression of PC markers, nuclear size, and nucleoli number, we confirmed that our method delivers a pure fraction of PC nuclei. To demonstrate its applicability, we isolated PC nuclei from spinocerebellar ataxia type 7 (SCA7) mice and identified transcriptional changes in known and new disease-associated genes. Access to pure PC nuclei offers insights into PC biology and pathology, including the nature of selective neuronal vulnerability.
Keywords: CP: Neuroscience; FANS; Purkinje cells; RanBP2; SCA7; cerebellum; isolation; neurodegeneration; nucleus; phosphodiesterase; spinocerebellar ataxia.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Cook A.A., Fields E., Watt A.J. Losing the Beat: Contribution of Purkinje Cell Firing Dysfunction to Disease, and Its Reversal. Neuroscience. 2021;462:247–261. - PubMed
-
- Tomomura M., Rice D.S., Morgan J.I., Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur. J. Neurosci. 2001;14:57–63. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

