Fibroblast-like synoviocytes orchestrate daily rhythmic inflammation in arthritis
- PMID: 38981514
- PMCID: PMC11285822
- DOI: 10.1098/rsob.240089
Fibroblast-like synoviocytes orchestrate daily rhythmic inflammation in arthritis
Abstract
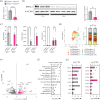
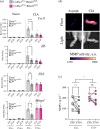
Rheumatoid arthritis is a chronic inflammatory disease that shows characteristic diurnal variation in symptom severity, where joint resident fibroblast-like synoviocytes (FLS) act as important mediators of arthritis pathology. We investigate the role of FLS circadian clock function in directing rhythmic joint inflammation in a murine model of inflammatory arthritis. We demonstrate FLS time-of-day-dependent gene expression is attenuated in arthritic joints, except for a subset of disease-modifying genes. The deletion of essential clock gene Bmal1 in FLS reduced susceptibility to collagen-induced arthritis but did not impact symptomatic severity in affected mice. Notably, FLS Bmal1 deletion resulted in loss of diurnal expression of disease-modulating genes across the joint, and elevated production of MMP3, a prognostic marker of joint damage in inflammatory arthritis. This work identifies the FLS circadian clock as an influential driver of daily oscillations in joint inflammation, and a potential regulator of destructive pathology in chronic inflammatory arthritis.
Keywords: circadian clock; fibroblast-like synoviocytes; inflammation; matrix metalloprotease; rheumatoid arthritis.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
