Start codon variant in LAG3 is associated with decreased LAG-3 expression and increased risk of autoimmune thyroid disease
- PMID: 38982041
- PMCID: PMC11233504
- DOI: 10.1038/s41467-024-50007-7
Start codon variant in LAG3 is associated with decreased LAG-3 expression and increased risk of autoimmune thyroid disease
Abstract
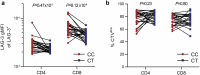
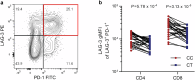
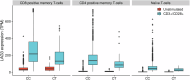
Autoimmune thyroid disease (AITD) is a common autoimmune disease. In a GWAS meta-analysis of 110,945 cases and 1,084,290 controls, 290 sequence variants at 225 loci are associated with AITD. Of these variants, 115 are previously unreported. Multiomics analysis yields 235 candidate genes outside the MHC-region and the findings highlight the importance of genes involved in T-cell regulation. A rare 5'-UTR variant (rs781745126-T, MAF = 0.13% in Iceland) in LAG3 has the largest effect (OR = 3.42, P = 2.2 × 10-16) and generates a novel start codon for an open reading frame upstream of the canonical protein translation initiation site. rs781745126-T reduces mRNA and surface expression of the inhibitory immune checkpoint LAG-3 co-receptor on activated lymphocyte subsets and halves LAG-3 levels in plasma among heterozygotes. All three homozygous carriers of rs781745126-T have AITD, of whom one also has two other T-cell mediated diseases, that is vitiligo and type 1 diabetes. rs781745126-T associates nominally with vitiligo (OR = 5.1, P = 6.5 × 10-3) but not with type 1 diabetes. Thus, the effect of rs781745126-T is akin to drugs that inhibit LAG-3, which unleash immune responses and can have thyroid dysfunction and vitiligo as adverse events. This illustrates how a multiomics approach can reveal potential drug targets and safety concerns.
© 2024. The Author(s).
Conflict of interest statement
S.S., K.B., T.M., J.B., T.A.O., G.H.H., G.R., K.G., A.O.A., S.H.L., L.S., J.G., A.S., A.O., B.V.H., E.F., E.V.I., G.S., G.M., G.H.E., G.A.T., K.K., K.H.S.M, S.A.G., S.R., H.H., O.T.M., P.S., D.F.G., T.R., G.T., P.M., G.L.N., I.J., and K.S. declare competing interests as employees of deCODE genetics/Amgen. The remaining authors declare no competing interests.
Figures


Similar articles
-
Type 1 Diabetes and Autoimmune Thyroid Disease-The Genetic Link.Front Endocrinol (Lausanne). 2021 Mar 10;12:618213. doi: 10.3389/fendo.2021.618213. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33776915 Free PMC article. Review.
-
Genome wide identification of new genes and pathways in patients with both autoimmune thyroiditis and type 1 diabetes.J Autoimmun. 2015 Jun;60:32-9. doi: 10.1016/j.jaut.2015.03.006. Epub 2015 Apr 27. J Autoimmun. 2015. PMID: 25936594 Free PMC article.
-
The association of CTLA4 polymorphism with type 1 diabetes is concentrated in patients complicated with autoimmune thyroid disease: a multicenter collaborative study in Japan.J Clin Endocrinol Metab. 2006 Mar;91(3):1087-92. doi: 10.1210/jc.2005-1407. Epub 2005 Dec 13. J Clin Endocrinol Metab. 2006. PMID: 16352685
-
The protein tyrosine phosphatase non-receptor type 22 C1858T polymorphism is a joint susceptibility locus for immunthyroiditis and autoimmune diabetes.Thyroid. 2009 Feb;19(2):143-8. doi: 10.1089/thy.2008.0301. Thyroid. 2009. PMID: 19090780
-
Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms.Endocr Rev. 2008 Oct;29(6):697-725. doi: 10.1210/er.2008-0015. Epub 2008 Sep 5. Endocr Rev. 2008. PMID: 18776148 Free PMC article. Review.
Cited by
-
From Phenotype to Molecules: Unveiling the Genetic and Immunological Bridges Between Autoimmune Diseases and Vitiligo.Clin Cosmet Investig Dermatol. 2024 Nov 4;17:2475-2486. doi: 10.2147/CCID.S488746. eCollection 2024. Clin Cosmet Investig Dermatol. 2024. PMID: 39524107 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

