Whole exome sequencing analysis identifies genes for alcohol consumption
- PMID: 38982111
- PMCID: PMC11233704
- DOI: 10.1038/s41467-024-50132-3
Whole exome sequencing analysis identifies genes for alcohol consumption
Abstract
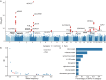
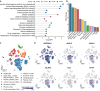
Alcohol consumption is a heritable behavior seriously endangers human health. However, genetic studies on alcohol consumption primarily focuses on common variants, while insights from rare coding variants are lacking. Here we leverage whole exome sequencing data across 304,119 white British individuals from UK Biobank to identify protein-coding variants associated with alcohol consumption. Twenty-five variants are associated with alcohol consumption through single variant analysis and thirteen genes through gene-based analysis, ten of which have not been reported previously. Notably, the two unreported alcohol consumption-related genes GIGYF1 and ANKRD12 show enrichment in brain function-related pathways including glial cell differentiation and are strongly expressed in the cerebellum. Phenome-wide association analyses reveal that alcohol consumption-related genes are associated with brain white matter integrity and risk of digestive and neuropsychiatric diseases. In summary, this study enhances the comprehension of the genetic architecture of alcohol consumption and implies biological mechanisms underlying alcohol-related adverse outcomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

