Identification of an H-Ras nanocluster disrupting peptide
- PMID: 38982284
- PMCID: PMC11233548
- DOI: 10.1038/s42003-024-06523-9
Identification of an H-Ras nanocluster disrupting peptide
Abstract
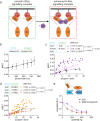
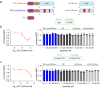
Hyperactive Ras signalling is found in most cancers. Ras proteins are only active in membrane nanoclusters, which are therefore potential drug targets. We previously showed that the nanocluster scaffold galectin-1 (Gal1) enhances H-Ras nanoclustering via direct interaction with the Ras binding domain (RBD) of Raf. Here, we establish that the B-Raf preference of Gal1 emerges from the divergence of the Raf RBDs at their proposed Gal1-binding interface. We then identify the L5UR peptide, which disrupts this interaction by binding with low micromolar affinity to the B- and C-Raf-RBDs. Its 23-mer core fragment is sufficient to interfere with H-Ras nanoclustering, modulate Ras-signalling and moderately reduce cell viability. These latter two phenotypic effects may also emerge from the ability of L5UR to broadly engage with several RBD- and RA-domain containing Ras interactors. The L5UR-peptide core fragment is a starting point for the development of more specific reagents against Ras-nanoclustering and -interactors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

