Protosappanin A Protects DOX-Induced Myocardial Injury and Cardiac Dysfunction by Targeting ACSL4/FTH1 Axis-Dependent Ferroptosis
- PMID: 38984448
- PMCID: PMC11425893
- DOI: 10.1002/advs.202310227
Protosappanin A Protects DOX-Induced Myocardial Injury and Cardiac Dysfunction by Targeting ACSL4/FTH1 Axis-Dependent Ferroptosis
Abstract
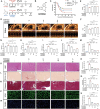
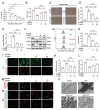
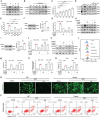
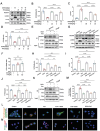
Doxorubicin (DOX) is an effective anticancer agent, but its clinical utility is constrained by dose-dependent cardiotoxicity, partly due to cardiomyocyte ferroptosis. However, the progress of developing cardioprotective medications to counteract ferroptosis has encountered obstacles. Protosappanin A (PrA), an anti-inflammatory compound derived from hematoxylin, shows potential against DOX-induced cardiomyopathy (DIC). Here, it is reported that PrA alleviates myocardial damage and dysfunction by reducing DOX-induced ferroptosis and maintaining mitochondrial homeostasis. Subsequently, the molecular target of PrA through proteome microarray, molecular docking, and dynamics simulation is identified. Mechanistically, PrA physically binds with ferroptosis-related proteins acyl-CoA synthetase long-chain family member 4 (ACSL4) and ferritin heavy chain 1 (FTH1), ultimately inhibiting ACSL4 phosphorylation and subsequent phospholipid peroxidation, while also preventing FTH1 autophagic degradation and subsequent release of ferrous ions (Fe2+) release. Given the critical role of ferroptosis in the pathogenesis of ischemia-reperfusion (IR) injury, this further investigation posits that PrA can confer a protective effect against IR-induced cardiac damage by inhibiting ferroptosis. Overall, a novel pharmacological inhibitor is unveiled that targets ferroptosis and uncover a dual-regulated mechanism for cardiomyocyte ferroptosis in DIC, highlighting additional therapeutic options for chemodrug-induced cardiotoxicity and ferroptosis-triggered disorders.
Keywords: doxorubicin‐induced cardiomyopathy; ferroptosis; molecular targeted therapy; myocardial injury; protosappanin A.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rayson D., Richel D., Chia S., Jackisch C., van der Vegt S., Suter T., Ann. Oncol. 2008, 19, 1530. - PubMed
-
- Raber I., Asnani A., Cardiovasc. Res. 2019, 115, 915. - PubMed
-
- Lyon A. R., Lopez‐Fernandez T., Couch L. S., Asteggiano R., Aznar M. C., Bergler‐Klein J., Boriani G., Cardinale D., Cordoba R., Cosyns B., Cutter D. J., de Azambuja E., de Boer R. A., Dent S. F., Farmakis D., Gevaert S. A., Gorog D. A., Herrmann J., Lenihan D., Moslehi J., Moura B., Salinger S. S., Stephens R., Suter T. M., Szmit S., Tamargo J., Thavendiranathan P., Tocchetti C. G., van der Meer P., van der Pal H. J. H., et al., Eur. Heart J. 2022, 43, 4229.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
