High-Resolution Mass Spectrometry for Human Exposomics: Expanding Chemical Space Coverage
- PMID: 38984754
- PMCID: PMC11271014
- DOI: 10.1021/acs.est.4c01156
High-Resolution Mass Spectrometry for Human Exposomics: Expanding Chemical Space Coverage
Abstract
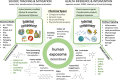
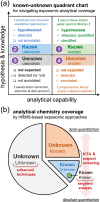
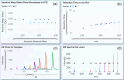
In the modern "omics" era, measurement of the human exposome is a critical missing link between genetic drivers and disease outcomes. High-resolution mass spectrometry (HRMS), routinely used in proteomics and metabolomics, has emerged as a leading technology to broadly profile chemical exposure agents and related biomolecules for accurate mass measurement, high sensitivity, rapid data acquisition, and increased resolution of chemical space. Non-targeted approaches are increasingly accessible, supporting a shift from conventional hypothesis-driven, quantitation-centric targeted analyses toward data-driven, hypothesis-generating chemical exposome-wide profiling. However, HRMS-based exposomics encounters unique challenges. New analytical and computational infrastructures are needed to expand the analysis coverage through streamlined, scalable, and harmonized workflows and data pipelines that permit longitudinal chemical exposome tracking, retrospective validation, and multi-omics integration for meaningful health-oriented inferences. In this article, we survey the literature on state-of-the-art HRMS-based technologies, review current analytical workflows and informatic pipelines, and provide an up-to-date reference on exposomic approaches for chemists, toxicologists, epidemiologists, care providers, and stakeholders in health sciences and medicine. We propose efforts to benchmark fit-for-purpose platforms for expanding coverage of chemical space, including gas/liquid chromatography-HRMS (GC-HRMS and LC-HRMS), and discuss opportunities, challenges, and strategies to advance the burgeoning field of the exposome.
Keywords: chemical space; chromatography; environmental exposures; exposome; high-resolution mass spectrometry; metabolomics; non-targeted analysis; toxicants.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Grants and funding
- U2C ES030163/ES/NIEHS NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- R01 AG067501/AG/NIA NIH HHS/United States
- RF1 AG066107/AG/NIA NIH HHS/United States
- R21 ES036033/ES/NIEHS NIH HHS/United States
- R25 GM143298/GM/NIGMS NIH HHS/United States
- R01 ES032831/ES/NIEHS NIH HHS/United States
- P42 ES031007/ES/NIEHS NIH HHS/United States
- P30 ES009089/ES/NIEHS NIH HHS/United States
- R03 OD034497/OD/NIH HHS/United States
- UL1 TR004419/TR/NCATS NIH HHS/United States
- P30 ES019776/ES/NIEHS NIH HHS/United States
- K01 ES035398/ES/NIEHS NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- R21 ES034187/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

