Mono-ADP-Ribosylation of Peptides: An Overview of Synthetic and Chemoenzymatic Methodologies
- PMID: 38984757
- PMCID: PMC11664928
- DOI: 10.1002/cbic.202400440
Mono-ADP-Ribosylation of Peptides: An Overview of Synthetic and Chemoenzymatic Methodologies
Abstract
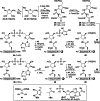
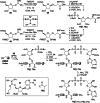
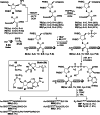
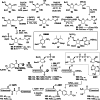
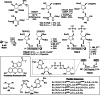
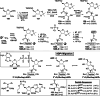
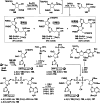
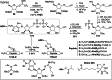
Adenosine diphosphate (ADP)-ribosylation is a ubiquitous post-translational modification that regulates vital biological processes like histone reorganization and DNA-damage repair through the modification of various amino acid residues. Due to advances in mass-spectrometry, the collection of long-known ADP-ribose (ADPr) acceptor sites, e. g. arginine, cysteine and glutamic acid, has been expanded with serine, tyrosine and histidine, among others. Well-defined ADPr-peptides are valuable tools for investigating the exact structures, mechanisms of action and interaction partners of the different flavors of this modification. This review provides a comprehensive overview of synthetic and chemoenzymatic methodologies that enabled the construction of peptides mono-ADP-ribosylated on various amino acids, and close mimetics thereof.
Keywords: chemoenzymatic synthesis; glycosylation; mono-ADP-ribosylated peptides; pyrophosphate; solid-phase peptide synthesis.
© 2024 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










Similar articles
-
Convergent Approach Toward ADP-Ribosylated Peptides via a Chemoselective Phosphate Condensation.Chemistry. 2025 Jul 8;31(38):e202501383. doi: 10.1002/chem.202501383. Epub 2025 Jun 16. Chemistry. 2025. PMID: 40471565 Free PMC article.
-
Chemical ADP-ribosylation: mono-ADPr-peptides and oligo-ADP-ribose.Org Biomol Chem. 2019 Jun 5;17(22):5460-5474. doi: 10.1039/c9ob00501c. Org Biomol Chem. 2019. PMID: 31112180 Review.
-
Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches.J Am Soc Mass Spectrom. 2021 Jan 6;32(1):157-168. doi: 10.1021/jasms.0c00040. Epub 2020 Nov 3. J Am Soc Mass Spectrom. 2021. PMID: 33140951
-
Molecular Tools for the Study of ADP-Ribosylation: A Unified and Versatile Method to Synthesise Native Mono-ADP-Ribosylated Peptides.Chemistry. 2021 Jul 21;27(41):10621-10627. doi: 10.1002/chem.202100337. Epub 2021 May 6. Chemistry. 2021. PMID: 33769608 Free PMC article.
-
Identification of ADP-ribosylated peptides and ADP-ribose acceptor sites.Front Biosci (Landmark Ed). 2014 Jun 1;19(7):1041-56. doi: 10.2741/4266. Front Biosci (Landmark Ed). 2014. PMID: 24896335 Review.
Cited by
-
Convergent Approach Toward ADP-Ribosylated Peptides via a Chemoselective Phosphate Condensation.Chemistry. 2025 Jul 8;31(38):e202501383. doi: 10.1002/chem.202501383. Epub 2025 Jun 16. Chemistry. 2025. PMID: 40471565 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

