Tumor-Intrinsic Activity of Chromobox 2 Remodels the Tumor Microenvironment in High-grade Serous Carcinoma
- PMID: 38984891
- PMCID: PMC11298703
- DOI: 10.1158/2767-9764.CRC-24-0027
Tumor-Intrinsic Activity of Chromobox 2 Remodels the Tumor Microenvironment in High-grade Serous Carcinoma
Abstract
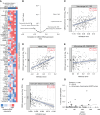
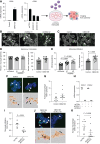
Chromobox 2 (CBX2), an epigenetic reader and component of polycomb repressor complex 1, is highly expressed in >75% of high-grade serous carcinoma. Increased CBX2 expression is associated with poorer survival, whereas CBX2 knockdown leads to improved chemotherapy sensitivity. In a high-grade serous carcinoma immune-competent murine model, knockdown of CBX2 decreased tumor progression. We sought to explore the impact of modulation of CBX2 on the tumor immune microenvironment (TIME), understanding that the TIME plays a critical role in disease progression and development of therapy resistance. Exploration of existing datasets demonstrated that elevated CBX2 expression significantly correlated with specific immune cell types in the TIME. RNA sequencing and pathway analysis of differentially expressed genes demonstrated immune signature enrichment. Confocal microscopy and co-culture experiments found that modulation of CBX2 leads to increased recruitment and infiltration of macrophages. Flow cytometry of macrophages cultured with CBX2-overexpressing cells showed increased M2-like macrophages and decreased phagocytosis activity. Cbx2 knockdown in the Trp53-null, Brca2-null ID8 syngeneic murine model (ID8 Trp53-/-Brca2-/-) led to decreased tumor progression compared with the control. NanoString immuno-oncology panel analysis suggested that knockdown in Cbx2 shifts immune cell composition, with an increase in macrophages. Multispectral immunohistochemistry (mIHC) further confirmed an increase in macrophage infiltration. Increased CBX2 expression leads to recruitment and polarization of protumor macrophages, and targeting CBX2 may serve to modulate the TIME to enhance the efficacy of immune therapies.
Significance: CBX2 expression correlates with the TIME. CBX2 modulation shifts the macrophage population, potentially leading to an immunosuppressive microenvironment, highlighting CBX2 as a target to improve efficacy of immunotherapy.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
T. Danhorn reports grants from NIH during the conduct of the study. B.G. Bitler reports grants from NCI/NIH, ACS, DOD, and OCRA during the conduct of the study. L.W. Brubaker reports grant from OCRA during the conduct of the study and consulting work with AstraZeneca outside the submitted work, as well as a patent to CBX2 inhibitor pending. No disclosures were reported by the other authors.
Figures



References
-
- Bowtell DDL. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer 2010;10:803–8. - PubMed
-
- McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet 2016;48:758–67. - PubMed
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. - PubMed
-
- Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet 2014;384:1376–88. - PubMed
-
- Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer 2003;3:502–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

