Active Site Characterization of a Campylobacter jejuni Nitrate Reductase Variant Provides Insight into the Enzyme Mechanism
- PMID: 38984973
- PMCID: PMC12088371
- DOI: 10.1021/acs.inorgchem.4c01991
Active Site Characterization of a Campylobacter jejuni Nitrate Reductase Variant Provides Insight into the Enzyme Mechanism
Abstract
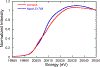
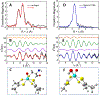
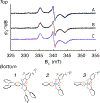
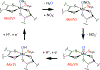
Mo K-edge X-ray absorption spectroscopy (XAS) is used to probe the structure of wild-type Campylobacter jejuni nitrate reductase NapA and the C176A variant. The results of extended X-ray absorption fine structure (EXAFS) experiments on wt NapA support an oxidized Mo(VI) hexacoordinate active site coordinated by a single terminal oxo donor, four sulfur atoms from two separate pyranopterin dithiolene ligands, and an additional S atom from a conserved cysteine amino acid residue. We found no evidence of a terminal sulfido ligand in wt NapA. EXAFS analysis shows the C176A active site to be a 6-coordinate structure, and this is supported by EPR studies on C176A and small molecule analogs of Mo(V) enzyme forms. The SCys is replaced by a hydroxide or water ligand in C176A, and we find no evidence of a coordinated sulfhydryl (SH) ligand. Kinetic studies show that this variant has completely lost its catalytic activity toward nitrate. Taken together, the results support a critical role for the conserved C176 in catalysis and an oxygen atom transfer mechanism for the catalytic reduction of nitrate to nitrite that does not employ a terminal sulfido ligand in the catalytic cycle.
Figures
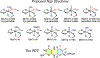
References
-
- Hille R; Schulzke C; Kirk ML Molybdenum and Tungsten Enzymes. In RSC Metallobiology Series, Garner CD, Sun H, Wedd A, Ciurli SL, Eds.; The Royal Society of Chemistry: Cambridge, UK, 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

