Dominant suppressor genes of p53-induced apoptosis in Drosophila melanogaster
- PMID: 38985658
- PMCID: PMC11373661
- DOI: 10.1093/g3journal/jkae149
Dominant suppressor genes of p53-induced apoptosis in Drosophila melanogaster
Abstract
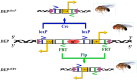
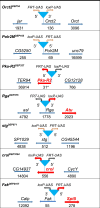
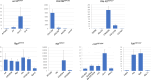
One of the major functions of programmed cell death (apoptosis) is the removal of cells that suffered oncogenic mutations, thereby preventing cancerous transformation. By making use of a Double-Headed-EP (DEP) transposon, a P element derivative made in our laboratory, we made an insertional mutagenesis screen in Drosophila melanogaster to identify genes that, when overexpressed, suppress the p53-activated apoptosis. The DEP element has Gal4-activatable, outward-directed UAS promoters at both ends, which can be deleted separately in vivo. In the DEP insertion mutants, we used the GMR-Gal4 driver to induce transcription from both UAS promoters and tested the suppression effect on the apoptotic rough eye phenotype generated by an activated UAS-p53 transgene. By DEP insertions, 7 genes were identified, which suppressed the p53-induced apoptosis. In 4 mutants, the suppression effect resulted from single genes activated by 1 UAS promoter (Pka-R2, Rga, crol, and Spt5). In the other 3 (Orct2, Polr2M, and stg), deleting either UAS promoter eliminated the suppression effect. In qPCR experiments, we found that the genes in the vicinity of the DEP insertion also showed an elevated expression level. This suggested an additive effect of the nearby genes on suppressing apoptosis. In the eukaryotic genomes, there are coexpressed gene clusters. Three of the DEP insertion mutants are included, and 2 are in close vicinity of separate coexpressed gene clusters. This raises the possibility that the activity of some of the genes in these clusters may help the suppression of the apoptotic cell death.
Keywords: Drosophila; activating insertional mutagenesis; apoptosis; p53; suppression.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures

References
-
- Aubrey BJ, Janic A, Chen Y, Chang C, Lieschke EC, Diepstraten ST, Kueh AJ, Bernardini JP, Dewson G, O'Reilly LA, et al. 2018a. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev. 32(21–22):1420–1429. doi: 10.1101/gad.314286.118. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
