Anti-hemagglutinin monomeric nanobody provides prophylactic immunity against H1 subtype influenza A viruses
- PMID: 38985719
- PMCID: PMC11236207
- DOI: 10.1371/journal.pone.0301664
Anti-hemagglutinin monomeric nanobody provides prophylactic immunity against H1 subtype influenza A viruses
Abstract
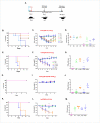
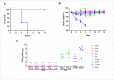
Influenza viruses constitute a major threat to human health globally. The viral surface glycoprotein hemagglutinin (HA) is the immunodominant antigen, contains the site for binding to the cellular receptor (RBS), and it is the major target of neutralizing antibody responses post-infection. We developed llama-derived single chain antibody fragments (VHHs) specific for type A influenza virus. Four VHHs were identified and further characterized. VHH D81 bound residues in the proximity of the C-terminal region of HA1 of H1 and H5 subtypes, and showed weak neutralizing activity, whereas VHH B33 bound residues in the proximity of the N-terminal region of the HA's stem domain (HA2) of H1, H5, and H9 subtypes, and showed no neutralizing activity. Of most relevance, VHHs E13 and G41 recognized highly conserved conformational epitopes on the H1 HA's globular domain (HA1) and showed high virus neutralizing activity (ranging between 0.94 to 0.01μM), when tested against several human H1N1 isolates. Additionally, E13 displayed abrogated virus replication of a panel of H1N1 strains spanning over 80 years of antigenic drift and isolated from human, avian, and swine origin. Interestingly, E13 conferred protection in vivo at a dose as low as 0.05 mg/kg. Mice treated with E13 intranasally resulted in undetectable virus challenge loads in the lungs at day 4 post-challenge. The transfer of sterilizing pan-H1 immunity, by a dose in the range of micrograms given intranasally, is of major significance for a monomeric VHH and supports the further development of E13 as an immunotherapeutic agent for the mitigation of influenza infections.
Copyright: © 2024 Barbieri et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- PAHO/OPS. PAHO/OPS | INFLUENZA REGIONAL UPDATE EW 5, 2022 /ACTUALIZACIÓN REGIONAL DE INFLUENZA SE 5 DE 2022. 2022.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

