Control of G protein-coupled receptor function via membrane-interacting intrinsically disordered C-terminal domains
- PMID: 38985766
- PMCID: PMC11260148
- DOI: 10.1073/pnas.2407744121
Control of G protein-coupled receptor function via membrane-interacting intrinsically disordered C-terminal domains
Abstract
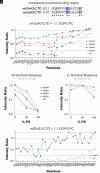
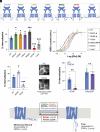
G protein-coupled receptors (GPCRs) control intracellular signaling cascades via agonist-dependent coupling to intracellular transducers including heterotrimeric G proteins, GPCR kinases (GRKs), and arrestins. In addition to their critical interactions with the transmembrane core of active GPCRs, all three classes of transducers have also been reported to interact with receptor C-terminal domains (CTDs). An underexplored aspect of GPCR CTDs is their possible role as lipid sensors given their proximity to the membrane. CTD-membrane interactions have the potential to control the accessibility of key regulatory CTD residues to downstream effectors and transducers. Here, we report that the CTDs of two closely related family C GPCRs, metabotropic glutamate receptor 2 (mGluR2) and mGluR3, bind to membranes and that this interaction can regulate receptor function. We first characterize CTD structure with NMR spectroscopy, revealing lipid composition-dependent modes of membrane binding. Using molecular dynamics simulations and structure-guided mutagenesis, we then identify key conserved residues and cancer-associated mutations that modulate CTD-membrane binding. Finally, we provide evidence that mGluR3 transducer coupling is controlled by CTD-membrane interactions in live cells, which may be subject to regulation by CTD phosphorylation and changes in membrane composition. This work reveals an additional mechanism of GPCR modulation, suggesting that CTD-membrane binding may be a general regulatory mode throughout the broad GPCR superfamily.
Keywords: GPCR; NMR; disordered protein; mGluR; β-arrestin.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




Update of
-
Control of G protein-coupled receptor function via membrane-interacting intrinsically disordered C-terminal domains.bioRxiv [Preprint]. 2024 May 2:2023.08.16.553551. doi: 10.1101/2023.08.16.553551. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2407744121. doi: 10.1073/pnas.2407744121. PMID: 37645938 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R35 GM136686/GM/NIGMS NIH HHS/United States
- F32GM148001/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R01NS129904/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- S10 OD028556/OD/NIH HHS/United States
- R35GM136686/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources

