Targeted genome editing restores auditory function in adult mice with progressive hearing loss caused by a human microRNA mutation
- PMID: 38985856
- PMCID: PMC7616320
- DOI: 10.1126/scitranslmed.adn0689
Targeted genome editing restores auditory function in adult mice with progressive hearing loss caused by a human microRNA mutation
Abstract
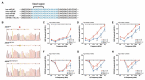
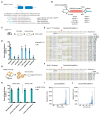
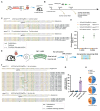
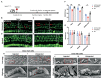
Mutations in microRNA-96 (MIR96) cause autosomal dominant deafness-50 (DFNA50), a form of delayed-onset hearing loss. Genome editing has shown efficacy in hearing recovery through intervention in neonatal mice, yet editing in the adult inner ear is necessary for clinical applications, which has not been done. Here, we developed a genome editing therapy for the MIR96 mutation 14C>A by screening different CRISPR systems and optimizing Cas9 expression and the sgRNA scaffold for efficient and specific mutation editing. AAV delivery of the KKH variant of Staphylococcus aureus Cas9 (SaCas9-KKH) and sgRNA to the cochleae of presymptomatic (3-week-old) and symptomatic (6-week-old) adult Mir9614C>A/+ mutant mice improved hearing long term, with efficacy increased by injection at a younger age. Adult inner ear delivery resulted in transient Cas9 expression without evidence of AAV genomic integration, indicating the good safety profile of our in vivo genome editing strategy. We developed a dual-AAV system, including an AAV-sgmiR96-master carrying sgRNAs against all known human MIR96 mutations. Because mouse and human MIR96 sequences share 100% homology, our approach and sgRNA selection for efficient and specific hair cell editing for long-term hearing recovery lay the foundation for the development of treatment for patients with DFNA50 caused by MIR96 mutations.
Conflict of interest statement
Figures



Update of
-
Targeted genome editing restores auditory function in adult mice with progressive hearing loss caused by a human microRNA mutation.bioRxiv [Preprint]. 2023 Oct 28:2023.10.26.564008. doi: 10.1101/2023.10.26.564008. bioRxiv. 2023. Update in: Sci Transl Med. 2024 Jul 10;16(755):eadn0689. doi: 10.1126/scitranslmed.adn0689. PMID: 37961137 Free PMC article. Updated. Preprint.
References
-
- Müller U, Barr-Gillespie PG. New treatment options for hearing loss. Nat Rev Drug Discov. 2015;14:346–365. - PubMed
-
- Mahmoudian-Sani MR, Mehri-Ghahfarrokhi A, Ahmadinejad F, Hashemzadeh-Chaleshtori M, Saidijam M, Jami M-S. MicroRNAs: Effective elements in ear-related diseases and hearing loss. Eur Arch Otorhinolaryngol. 2017;274:2373–2380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

