Thermodynamic profiles for cotranslational trigger factor substrate recognition
- PMID: 38985872
- PMCID: PMC11235164
- DOI: 10.1126/sciadv.adn4824
Thermodynamic profiles for cotranslational trigger factor substrate recognition
Abstract
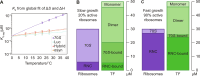
Molecular chaperones are central to the maintenance of proteostasis in living cells. A key member of this protein family is trigger factor (TF), which acts throughout the protein life cycle and has a ubiquitous role as the first chaperone encountered by proteins during synthesis. However, our understanding of how TF achieves favorable interactions with such a diverse substrate base remains limited. Here, we use microfluidics to reveal the thermodynamic determinants of this process. We find that TF binding to empty 70S ribosomes is enthalpy-driven, with micromolar affinity, while nanomolar affinity is achieved through a favorable entropic contribution for both intrinsically disordered and folding-competent nascent chains. These findings suggest a general mechanism for cotranslational TF function, which relies on occupation of the exposed TF-substrate binding groove rather than specific complementarity between chaperone and nascent chain. These insights add to our wider understanding of how proteins can achieve broad substrate specificity.
Figures


References
-
- Agashe V. R., Guha S., Chang H. C., Genevaux P., Hayer-Hartl M., Stemp M., Georgopoulos C., Hartl F. U., Barral J. M., Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell 117, 199–209 (2004). - PubMed
-
- Ferbitz L., Maier T., Patzelt H., Bukau B., Deuerling E., Ban N., Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004). - PubMed
-
- Kaiser C. M., Chang H. C., Agashe V. R., Lakshmipathy S. K., Etchells S. A., Hayer-Hartl M., Hartl F. U., Barral J. M., Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 (2006). - PubMed
-
- Rutkowska A., Mayer M. P., Hoffmann A., Merz F., Zachmann-Brand B., Schaffitzel C., Ban N., Deuerling E., Bukau B., Dynamics of trigger factor interaction with translating ribosomes. J. Biol. Chem. 283, 4124–4132 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

