Monoallelic de novo AJAP1 loss-of-function variants disrupt trans-synaptic control of neurotransmitter release
- PMID: 38985877
- PMCID: PMC11235169
- DOI: 10.1126/sciadv.adk5462
Monoallelic de novo AJAP1 loss-of-function variants disrupt trans-synaptic control of neurotransmitter release
Abstract
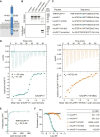
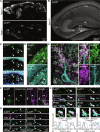
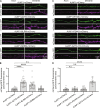
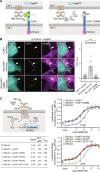
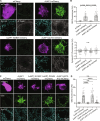
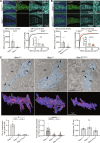
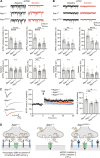
Adherens junction-associated protein 1 (AJAP1) has been implicated in brain diseases; however, a pathogenic mechanism has not been identified. AJAP1 is widely expressed in neurons and binds to γ-aminobutyric acid type B receptors (GBRs), which inhibit neurotransmitter release at most synapses in the brain. Here, we show that AJAP1 is selectively expressed in dendrites and trans-synaptically recruits GBRs to presynaptic sites of neurons expressing AJAP1. We have identified several monoallelic AJAP1 variants in individuals with epilepsy and/or neurodevelopmental disorders. Specifically, we show that the variant p.(W183C) lacks binding to GBRs, resulting in the inability to recruit them. Ultrastructural analysis revealed significantly decreased presynaptic GBR levels in Ajap1-/- and Ajap1W183C/+ mice. Consequently, these mice exhibited reduced GBR-mediated presynaptic inhibition at excitatory and inhibitory synapses, along with impaired synaptic plasticity. Our study reveals that AJAP1 enables the postsynaptic neuron to regulate the level of presynaptic GBR-mediated inhibition, supporting the clinical relevance of loss-of-function AJAP1 variants.
Figures
References
-
- Anttila V., Winsvold B. S., Gormley P., Kurth T., Bettella F., McMahon G., Kallela M., Malik R., de Vries B., Terwindt G., Medland S. E., Todt U., McArdle W. L., Quaye L., Koiranen M., Ikram M. A., Lehtimaki T., Stam A. H., Ligthart L., Wedenoja J., Dunham I., Neale B. M., Palta P., Hamalainen E., Schurks M., Rose L. M., Buring J. E., Ridker P. M., Steinberg S., Stefansson H., Jakobsson F., Lawlor D. A., Evans D. M., Ring S. M., Farkkila M., Artto V., Kaunisto M. A., Freilinger T., Schoenen J., Frants R. R., Pelzer N., Weller C. M., Zielman R., Heath A. C., Madden P. A. F., Montgomery G. W., Martin N. G., Borck G., Gobel H., Heinze A., Heinze-Kuhn K., Williams F. M. K., Hartikainen A. L., Pouta A., van den Ende J., Uitterlinden A. G., Hofman A., Amin N., Hottenga J. J., Vink J. M., Heikkila K., Alexander M., Muller-Myhsok B., Schreiber S., Meitinger T., Wichmann H. E., Aromaa A., Eriksson J. G., Traynor B., Trabzuni D., North American Brain Expression Consortium, UK Brain Expression Consortium, Rossin E., Lage K., Jacobs S. B. R., Gibbs J. R., Birney E., Kaprio J., Penninx B. W., Boomsma D. I., van Duijn C., Raitakari O., Jarvelin M. R., Zwart J. A., Cherkas L., Strachan D. P., Kubisch C., Ferrari M. D., van den Maagdenberg A., Dichgans M., Wessman M., Smith G. D., Stefansson K., Daly M. J., Nyholt D. R., Chasman D., Palotie A., Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat. Genet. 45, 912–917 (2013). - PMC - PubMed
-
- Zhang M., Zhou X., Jiang W., Li M., Zhou R., Zhou S., AJAP1 affects behavioral changes and GABABR1 level in epileptic mice. Biochem. Biophys. Res. Commun. 524, 1057–1063 (2020). - PubMed
-
- Ikeda M., Tomita Y., Mouri A., Koga M., Okochi T., Yoshimura R., Yamanouchi Y., Kinoshita Y., Hashimoto R., Williams H. J., Takeda M., Nakamura J., Nabeshima T., Owen M. J., O'Donovan M. C., Honda H., Arinami T., Ozaki N., Iwata N., Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: Integrated analysis among pharmacogenomics, mouse expression, and genetic case-control association approaches. Biol. Psychiatry 67, 263–269 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

