Requirement of Pdgfrα+ cells for calvarial bone repair
- PMID: 38986535
- PMCID: PMC11328938
- DOI: 10.1093/stcltm/szae041
Requirement of Pdgfrα+ cells for calvarial bone repair
Abstract
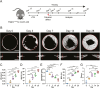
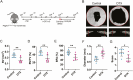
Platelet-derived growth factor receptor α (PDGFRα) is often considered as a general marker of mesenchymal cells and fibroblasts, but also shows expression in a portion of osteoprogenitor cells. Within the skeleton, Pdgfrα+ mesenchymal cells have been identified in bone marrow and periosteum of long bones, where they play a crucial role in participating in fracture repair. A similar examination of Pdgfrα+ cells in calvarial bone healing has not been examined. Here, we utilize Pdgfrα-CreERTM;mT/mG reporter animals to examine the contribution of Pdgfrα+ mesenchymal cells to calvarial bone repair through histology and single-cell RNA sequencing (scRNA-Seq). Results showed that Pdgfrα+ mesenchymal cells are present in several cell clusters by scRNA-Seq, and by histology a dramatic increase in Pdgfrα+ cells populated the defect site at early timepoints to give rise to healed bone tissue overtime. Notably, diphtheria toxin-mediated ablation of Pdgfrα reporter+ cells resulted in significantly impaired calvarial bone healing. Our findings suggest that Pdgfrα-expressing cells within the calvarial niche play a critical role in the process of calvarial bone repair.
Keywords: bone healing; bone repair; calvarial bone; mesenchymal cells; platelet-derived growth factor receptor; single-cell RNA sequencing; skeletal cells.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
A.W.J. declared scientific advisory board for Novadip LLC, consultant for Lifesprout LLC and Novadip LLC, and Editorial Board of Bone Research, Stem Cells, and The American Journal of Pathology. All the other authors declared no potential conflicts of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

