RNA biogenesis and RNA metabolism factors as R-loop suppressors: a hidden role in genome integrity
- PMID: 38986581
- PMCID: PMC11293400
- DOI: 10.1101/gad.351853.124
RNA biogenesis and RNA metabolism factors as R-loop suppressors: a hidden role in genome integrity
Abstract
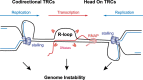
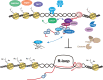
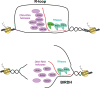
Genome integrity relies on the accuracy of DNA metabolism, but as appreciated for more than four decades, transcription enhances mutation and recombination frequencies. More recent research provided evidence for a previously unforeseen link between RNA and DNA metabolism, which is often related to the accumulation of DNA-RNA hybrids and R-loops. In addition to physiological roles, R-loops interfere with DNA replication and repair, providing a molecular scenario for the origin of genome instability. Here, we review current knowledge on the multiple RNA factors that prevent or resolve R-loops and consequent transcription-replication conflicts and thus act as modulators of genome dynamics.
Keywords: DNA–RNA hybrids; R-loops; RNA biogenesis; RNA metabolism; genome instability; helicases.
© 2024 Luna et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
