Disruption of TGF-β signaling pathway is required to mediate effective killing of hepatocellular carcinoma by human iPSC-derived NK cells
- PMID: 38986609
- PMCID: PMC11380586
- DOI: 10.1016/j.stem.2024.06.009
Disruption of TGF-β signaling pathway is required to mediate effective killing of hepatocellular carcinoma by human iPSC-derived NK cells
Abstract
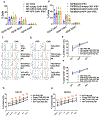
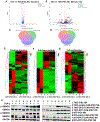
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. Transforming growth factor beta (TGF-β) is highly expressed in the liver tumor microenvironment and is known to inhibit immune cell activity. Here, we used human induced pluripotent stem cells (iPSCs) to produce natural killer (NK) cells engineered to mediate improved anti-HCC activity. Specifically, we produced iPSC-NK cells with either knockout TGF-β receptor 2 (TGFBR2-KO) or expression of a dominant negative (DN) form of the TGF-β receptor 2 (TGFBR2-DN) combined with chimeric antigen receptors (CARs) that target either GPC3 or AFP. The TGFBR2-KO and TGFBR2-DN iPSC-NK cells are resistant to TGF-β inhibition and improved anti-HCC activity. However, expression of anti-HCC CARs on iPSC-NK cells did not lead to effective anti-HCC activity unless there was also inhibition of TGF-β activity. Our findings demonstrate that TGF-β signaling blockade is required for effective NK cell function against HCC and potentially other malignancies that express high levels of TGF-β.
Keywords: AFP; GPC3; TGF-β signaling; alpha-fetoprotein and CAR-NK cells; glypican 3; hepatocellular carcinoma; iPSC-NK cells; tumor microenvironment.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.S.K. is a co-founder and advisor to Shoreline Biosciences and has an equity interest in the company. D.S.K. also consults VisiCELL Medical and RedC Bio for which he receives income and/or equity. Studies in this work are not related to the work of those companies. The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies.
Figures





References
-
- Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F, Wang Q, Wang S, Rong D, Reiter FP, et al. (2020). The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduction and Targeted Therapy 5, 87. 10.1038/s41392-020-0187-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

