Chimeric antigen receptor-induced antigen loss protects CD5.CART cells from fratricide without compromising on-target cytotoxicity
- PMID: 38986621
- PMCID: PMC11293353
- DOI: 10.1016/j.xcrm.2024.101628
Chimeric antigen receptor-induced antigen loss protects CD5.CART cells from fratricide without compromising on-target cytotoxicity
Abstract
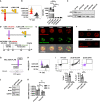
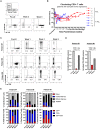
Chimeric antigen receptor T cells (CART) targeting lymphocyte antigens can induce T cell fratricide and require additional engineering to mitigate self-damage. We demonstrate that the expression of a chimeric antigen receptor (CAR) targeting CD5, a prominent pan-T cell antigen, induces rapid internalization and complete loss of the CD5 protein on T cells, protecting them from self-targeting. Notably, exposure of healthy and malignant T cells to CD5.CART cells induces similar internalization of CD5 on target cells, transiently shielding them from cytotoxicity. However, this protection is short-lived, as sustained activity of CD5.CART cells in patients with T cell malignancies results in full ablation of CD5+ T cells while sparing healthy T cells naturally lacking CD5. These results indicate that continuous downmodulation of the target antigen in CD5.CART cells produces effective fratricide resistance without undermining their on-target cytotoxicity.
Keywords: CAR; CD5; T cell fratricide; T cell malignancies; chimeric antigen receptor; clinical trials; fratricide; off-tumor toxicity.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.M. and M.K.B. are co-founders of March Biosciences, have equity, and serve on the advisory board for March Biosciences. M.K.B. is a cofounder and equity holder in AlloVir, Inc. and Marker Therapeutics. M.K.B. has equity in Tessa Therapeutics Ltd. and March Biosciences and serves on advisory boards for Marker Therapeutics, Allogene, Walking Fish, Abintus, Tessa Therapeutics, Athenex, ONK Therapeutics, Coya Therapeutics, Triumvira, Adaptimmune, Vor Therapeutics, and TScan. M.M. serves on the scientific advisory board of NKILT and receives research support from Fate Therapeutics and honoraria from Amgen and Galapagos NV. L.C.H. provides consulting services to March Biosciences. M.M. and M.K.B. have patent applications and a granted patent related to this work. An up-to-date declaration of interests for the listed authors can be found at https://www.bcm.edu/academic-centers/cell-and-gene-therapy/research/disclosure-of-outside-interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

