Ribonuclease 4 functions as an intestinal antimicrobial protein to maintain gut microbiota and metabolite homeostasis
- PMID: 38987259
- PMCID: PMC11237007
- DOI: 10.1038/s41467-024-50223-1
Ribonuclease 4 functions as an intestinal antimicrobial protein to maintain gut microbiota and metabolite homeostasis
Abstract
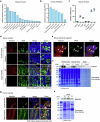
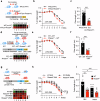
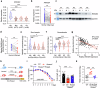
Antimicrobial proteins contribute to host-microbiota interactions and are associated with inflammatory bowel disease (IBD), but our understanding on antimicrobial protein diversity and functions remains incomplete. Ribonuclease 4 (Rnase4) is a potential antimicrobial protein with no known function in the intestines. Here we find that RNASE4 is expressed in intestinal epithelial cells (IEC) including Paneth and goblet cells, and is detectable in human and mouse stool. Results from Rnase4-deficient mice and recombinant protein suggest that Rnase4 kills Parasutterella to modulate intestinal microbiome, thereby enhancing indoleamine-2,3-dioxygenase 1 (IDO1) expression and subsequently kynurenic and xanthurenic acid production in IECs to reduce colitis susceptibility. Furthermore, deceased RNASE4 levels are observed in the intestinal tissues and stool from patients with IBD, correlating with increased stool Parasutterella. Our results thus implicate Rnase4 as an intestinal antimicrobial protein regulating gut microbiota and metabolite homeostasis, and as a potential diagnostic biomarker and therapeutic target for IBD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

