PARG is essential for Polθ-mediated DNA end-joining by removing repressive poly-ADP-ribose marks
- PMID: 38987289
- PMCID: PMC11236980
- DOI: 10.1038/s41467-024-50158-7
PARG is essential for Polθ-mediated DNA end-joining by removing repressive poly-ADP-ribose marks
Abstract
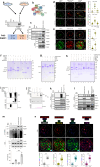
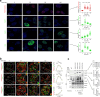
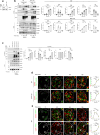
DNA polymerase theta (Polθ)-mediated end-joining (TMEJ) repairs DNA double-strand breaks and confers resistance to genotoxic agents. How Polθ is regulated at the molecular level to exert TMEJ remains poorly characterized. We find that Polθ interacts with and is PARylated by PARP1 in a HPF1-independent manner. PARP1 recruits Polθ to the vicinity of DNA damage via PARylation dependent liquid demixing, however, PARylated Polθ cannot perform TMEJ due to its inability to bind DNA. PARG-mediated de-PARylation of Polθ reactivates its DNA binding and end-joining activities. Consistent with this, PARG is essential for TMEJ and the temporal recruitment of PARG to DNA damage corresponds with TMEJ activation and dissipation of PARP1 and PAR. In conclusion, we show a two-step spatiotemporal mechanism of TMEJ regulation. First, PARP1 PARylates Polθ and facilitates its recruitment to DNA damage sites in an inactivated state. PARG subsequently activates TMEJ by removing repressive PAR marks on Polθ.
© 2024. The Author(s).
Conflict of interest statement
R.T.P. is a co-founder and chief scientific officer of Recombination Therapeutics, LLC. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- R01CA244707/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01GM137124/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01CA244179/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA283396/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R35 GM152198/GM/NIGMS NIH HHS/United States
- R01 CA283396/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- R50 CA221838/CA/NCI NIH HHS/United States
- R01 GM137124/GM/NIGMS NIH HHS/United States
- TRP 6628-21/Leukemia and Lymphoma Society (Leukemia & Lymphoma Society)
- R01 CA247707/CA/NCI NIH HHS/United States
- R01CA186238/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA186238/CA/NCI NIH HHS/United States
- R01 CA244179/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

