Early mutational signatures and transmissibility of SARS-CoV-2 Gamma and Lambda variants in Chile
- PMID: 38987406
- PMCID: PMC11237036
- DOI: 10.1038/s41598-024-66885-2
Early mutational signatures and transmissibility of SARS-CoV-2 Gamma and Lambda variants in Chile
Abstract
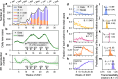
Genomic surveillance (GS) programmes were crucial in identifying and quantifying the mutating patterns of SARS-CoV-2 during the COVID-19 pandemic. In this work, we develop a Bayesian framework to quantify the relative transmissibility of different variants tailored for regions with limited GS. We use it to study the relative transmissibility of SARS-CoV-2 variants in Chile. Among the 3443 SARS-CoV-2 genomes collected between January and June 2021, where sampling was designed to be representative, the Gamma (P.1), Lambda (C.37), Alpha (B.1.1.7), B.1.1.348, and B.1.1 lineages were predominant. We found that Lambda and Gamma variants' reproduction numbers were 5% (95% CI: [1%, 14%]) and 16% (95% CI: [11%, 21%]) larger than Alpha's, respectively. Besides, we observed a systematic mutation enrichment in the Spike gene for all circulating variants, which strongly correlated with variants' transmissibility during the studied period (r = 0.93, p-value = 0.025). We also characterised the mutational signatures of local samples and their evolution over time and with the progress of vaccination, comparing them with those of samples collected in other regions worldwide. Altogether, our work provides a reliable method for quantifying variant transmissibility under subsampling and emphasises the importance of continuous genomic surveillance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Van Egeren D, et al. Risk of evolutionary escape from neutralizing antibodies targeting SARS-CoV-2 spike protein. MedRxiv. 2020;2020:11.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

