Dataset of the rumen microbiota and epithelial transcriptomics and proteomics in goat affected by solid diets
- PMID: 38987532
- PMCID: PMC11237100
- DOI: 10.1038/s41597-024-03584-7
Dataset of the rumen microbiota and epithelial transcriptomics and proteomics in goat affected by solid diets
Abstract
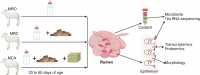
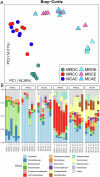
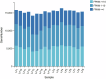
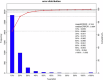
Although early solid diet supplementation is a common practice to improve the growth and development in goat kids, its biological mechanism how solid diet induces rumen microbiota and epithelial development is still unknow. In this study, rumen fermentation parameters, 16S rRNA sequencing for rumen content and epithelial microbiota, transcriptomics and proteomics of epithelium were determined to classify the effects of solid diet supplementation. Here, we classified the changes of goat phenotypes (i.e., growth performance, rumen fermentation and development) and linked them to the changes of rumen microbiota, transcriptome and expressed proteins. The mechanism of solid diet improving rumen development was elucidated preliminarily. Moreover, different roles between the rumen content and epithelial microbiota were identified. Thess datasets expands our understanding of the association between the early diet intervention and rumen development, providing the useful information how nutrient strategy affects rumen function and subsequently improves the host growth. The generated data provides insights in the importance of rumen niche microbiota and microbe-host interactions, which benefits future studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats.Microbiome. 2017 Sep 19;5(1):123. doi: 10.1186/s40168-017-0341-z. Microbiome. 2017. PMID: 28927467 Free PMC article.
-
Solid diet manipulates rumen epithelial microbiota and its interactions with host transcriptomic in young ruminants.Environ Microbiol. 2021 Nov;23(11):6557-6568. doi: 10.1111/1462-2920.15757. Epub 2021 Sep 20. Environ Microbiol. 2021. PMID: 34490978 Free PMC article.
-
Corn oil supplementation enhances hydrogen use for biohydrogenation, inhibits methanogenesis, and alters fermentation pathways and the microbial community in the rumen of goats.J Anim Sci. 2019 Dec 17;97(12):4999-5008. doi: 10.1093/jas/skz352. J Anim Sci. 2019. PMID: 31740932 Free PMC article.
-
Comparative aspects of plant tannins on digestive physiology, nutrition and microbial community changes in sheep and goats: A review.J Anim Physiol Anim Nutr (Berl). 2018 Oct;102(5):1181-1193. doi: 10.1111/jpn.12938. Epub 2018 Jul 24. J Anim Physiol Anim Nutr (Berl). 2018. PMID: 30039875 Review.
-
Development and physiology of the rumen and the lower gut: Targets for improving gut health.J Dairy Sci. 2016 Jun;99(6):4955-4966. doi: 10.3168/jds.2015-10351. Epub 2016 Mar 9. J Dairy Sci. 2016. PMID: 26971143 Review.
Cited by
-
Transcriptomic characterization of the functional and morphological development of the rumen wall in weaned lambs fed a diet containing yeast co-cultures of Saccharomyces cerevisiae and Kluyveromyces marxianus.Front Vet Sci. 2025 Jan 22;12:1510689. doi: 10.3389/fvets.2025.1510689. eCollection 2025. Front Vet Sci. 2025. PMID: 39911691 Free PMC article.
-
Chestnut tannins impair ruminal fiber degradation through modulation of cellulolytic bacterial activity in sheep: an in vivo and in vitro investigation.J Anim Sci. 2025 Jan 4;103:skaf189. doi: 10.1093/jas/skaf189. J Anim Sci. 2025. PMID: 40457894
-
Transcriptomics data for muscle development in Goats.Sci Data. 2025 Jun 2;12(1):928. doi: 10.1038/s41597-025-04950-9. Sci Data. 2025. PMID: 40456801 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

