The Hippo pathway transcription factors YAP and TAZ play HPV-type dependent roles in cervical cancer
- PMID: 38987584
- PMCID: PMC11237029
- DOI: 10.1038/s41467-024-49965-9
The Hippo pathway transcription factors YAP and TAZ play HPV-type dependent roles in cervical cancer
Abstract
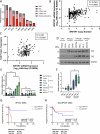
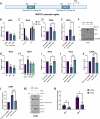
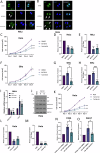
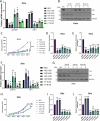
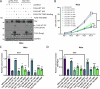
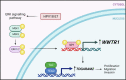
Human papillomaviruses (HPVs) cause most cervical cancers and an increasing number of anogenital and oral carcinomas, with most cases caused by HPV16 or HPV18. HPV hijacks host signalling pathways to promote carcinogenesis. Understanding these interactions could permit identification of much-needed therapeutics for HPV-driven malignancies. The Hippo signalling pathway is important in HPV+ cancers, with the downstream effector YAP playing a pro-oncogenic role. In contrast, the significance of its paralogue TAZ remains largely uncharacterised in these cancers. We demonstrate that TAZ is dysregulated in a HPV-type dependent manner by a distinct mechanism to that of YAP and controls proliferation via alternative cellular targets. Analysis of cervical cancer cell lines and patient biopsies revealed that TAZ expression was only significantly increased in HPV18+ and HPV18-like cells and TAZ knockdown reduced proliferation, migration and invasion only in HPV18+ cells. RNA-sequencing of HPV18+ cervical cells revealed that YAP and TAZ have distinct targets, suggesting they promote carcinogenesis by different mechanisms. Thus, in HPV18+ cancers, YAP and TAZ play non-redundant roles. This analysis identified TOGARAM2 as a previously uncharacterised TAZ target and demonstrates its role as a key effector of TAZ-mediated proliferation, migration and invasion in HPV18+ cancers.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

