Brainstem Dbh+ neurons control allergen-induced airway hyperreactivity
- PMID: 38987587
- PMCID: PMC11254774
- DOI: 10.1038/s41586-024-07608-5
Brainstem Dbh+ neurons control allergen-induced airway hyperreactivity
Abstract
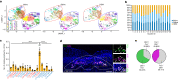
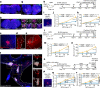
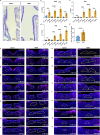
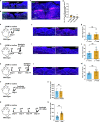
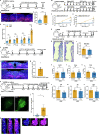
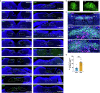
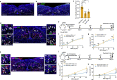
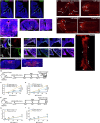
Exaggerated airway constriction triggered by repeated exposure to allergen, also called hyperreactivity, is a hallmark of asthma. Whereas vagal sensory neurons are known to function in allergen-induced hyperreactivity1-3, the identity of downstream nodes remains poorly understood. Here we mapped a full allergen circuit from the lung to the brainstem and back to the lung. Repeated exposure of mice to inhaled allergen activated the nuclei of solitary tract (nTS) neurons in a mast cell-, interleukin-4 (IL-4)- and vagal nerve-dependent manner. Single-nucleus RNA sequencing, followed by RNAscope assay at baseline and allergen challenges, showed that a Dbh+ nTS population is preferentially activated. Ablation or chemogenetic inactivation of Dbh+ nTS neurons blunted hyperreactivity whereas chemogenetic activation promoted it. Viral tracing indicated that Dbh+ nTS neurons project to the nucleus ambiguus (NA) and that NA neurons are necessary and sufficient to relay allergen signals to postganglionic neurons that directly drive airway constriction. Delivery of noradrenaline antagonists to the NA blunted hyperreactivity, suggesting noradrenaline as the transmitter between Dbh+ nTS and NA. Together, these findings provide molecular, anatomical and functional definitions of key nodes of a canonical allergen response circuit. This knowledge informs how neural modulation could be used to control allergen-induced airway hyperreactivity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Update of
-
Brainstem Dbh+ Neurons Control Chronic Allergen-Induced Airway Hyperreactivity.bioRxiv [Preprint]. 2024 Feb 14:2023.02.04.527145. doi: 10.1101/2023.02.04.527145. bioRxiv. 2024. Update in: Nature. 2024 Jul;631(8021):601-609. doi: 10.1038/s41586-024-07608-5. PMID: 36778350 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

