A liver immune rheostat regulates CD8 T cell immunity in chronic HBV infection
- PMID: 38987588
- PMCID: PMC11269190
- DOI: 10.1038/s41586-024-07630-7
A liver immune rheostat regulates CD8 T cell immunity in chronic HBV infection
Abstract
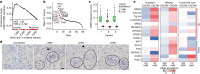
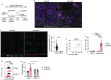
Chronic hepatitis B virus (HBV) infection affects 300 million patients worldwide1,2, in whom virus-specific CD8 T cells by still ill-defined mechanisms lose their function and cannot eliminate HBV-infected hepatocytes3-7. Here we demonstrate that a liver immune rheostat renders virus-specific CD8 T cells refractory to activation and leads to their loss of effector functions. In preclinical models of persistent infection with hepatotropic viruses such as HBV, dysfunctional virus-specific CXCR6+ CD8 T cells accumulated in the liver and, as a characteristic hallmark, showed enhanced transcriptional activity of cAMP-responsive element modulator (CREM) distinct from T cell exhaustion. In patients with chronic hepatitis B, circulating and intrahepatic HBV-specific CXCR6+ CD8 T cells with enhanced CREM expression and transcriptional activity were detected at a frequency of 12-22% of HBV-specific CD8 T cells. Knocking out the inhibitory CREM/ICER isoform in T cells, however, failed to rescue T cell immunity. This indicates that CREM activity was a consequence, rather than the cause, of loss in T cell function, further supported by the observation of enhanced phosphorylation of protein kinase A (PKA) which is upstream of CREM. Indeed, we found that enhanced cAMP-PKA-signalling from increased T cell adenylyl cyclase activity augmented CREM activity and curbed T cell activation and effector function in persistent hepatic infection. Mechanistically, CD8 T cells recognizing their antigen on hepatocytes established close and extensive contact with liver sinusoidal endothelial cells, thereby enhancing adenylyl cyclase-cAMP-PKA signalling in T cells. In these hepatic CD8 T cells, which recognize their antigen on hepatocytes, phosphorylation of key signalling kinases of the T cell receptor signalling pathway was impaired, which rendered them refractory to activation. Thus, close contact with liver sinusoidal endothelial cells curbs the activation and effector function of HBV-specific CD8 T cells that target hepatocytes expressing viral antigens by means of the adenylyl cyclase-cAMP-PKA axis in an immune rheostat-like fashion.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Wiktor, S. Z. & Hutin, Y. J. F. The global burden of viral hepatitis: better estimates to guide hepatitis elimination efforts. Lancet388, 1030–1031 (2016). - PubMed
-
- Thomas, D. L. Global elimination of chronic hepatitis. New Engl. J. Med.380, 2041–2050 (2019). - PubMed
-
- Fisicaro, P. et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med.23, 327–336 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

