Single-cell atlas of the human brain vasculature across development, adulthood and disease
- PMID: 38987604
- PMCID: PMC11324530
- DOI: 10.1038/s41586-024-07493-y
Single-cell atlas of the human brain vasculature across development, adulthood and disease
Abstract
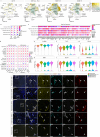
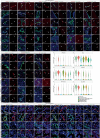
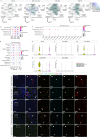
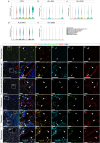
A broad range of brain pathologies critically relies on the vasculature, and cerebrovascular disease is a leading cause of death worldwide. However, the cellular and molecular architecture of the human brain vasculature remains incompletely understood1. Here we performed single-cell RNA sequencing analysis of 606,380 freshly isolated endothelial cells, perivascular cells and other tissue-derived cells from 117 samples, from 68 human fetuses and adult patients to construct a molecular atlas of the developing fetal, adult control and diseased human brain vasculature. We identify extensive molecular heterogeneity of the vasculature of healthy fetal and adult human brains and across five vascular-dependent central nervous system (CNS) pathologies, including brain tumours and brain vascular malformations. We identify alteration of arteriovenous differentiation and reactivated fetal as well as conserved dysregulated genes and pathways in the diseased vasculature. Pathological endothelial cells display a loss of CNS-specific properties and reveal an upregulation of MHC class II molecules, indicating atypical features of CNS endothelial cells. Cell-cell interaction analyses predict substantial endothelial-to-perivascular cell ligand-receptor cross-talk, including immune-related and angiogenic pathways, thereby revealing a central role for the endothelium within brain neurovascular unit signalling networks. Our single-cell brain atlas provides insights into the molecular architecture and heterogeneity of the developing, adult/control and diseased human brain vasculature and serves as a powerful reference for future studies.
© 2024. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures














Comment in
-
The molecular landscape of vascular cells in the human brain.Nat Rev Cardiol. 2024 Dec;21(12):847-848. doi: 10.1038/s41569-024-01079-x. Nat Rev Cardiol. 2024. PMID: 39304749 Free PMC article.
References
-
- Cho, C., Smallwood, P. M. & Nathans, J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-Specific signaling in mammalian CNS angiogenesis and blood-brain barrier regulation. Neuron95, 1221–1225 (2017). - PubMed
-
- Wälchli, T. et al. Wiring the vascular network with neural cues: a CNS perspective. Neuron87, 271–296 (2015). - PubMed
-
- Zlokovic, B. V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron57, 178–201 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

