High-sensitivity whole-mount in situ Hybridization of Mouse Oocytes and Embryos Visualizes the Super-resolution Structures and Distributions of mRNA Molecules
- PMID: 38987687
- PMCID: PMC11234658
- DOI: 10.1186/s12575-024-00250-5
High-sensitivity whole-mount in situ Hybridization of Mouse Oocytes and Embryos Visualizes the Super-resolution Structures and Distributions of mRNA Molecules
Abstract
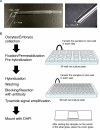
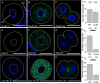
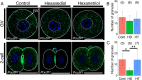
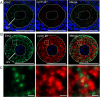
Mammalian oocytes accumulate more than ten thousand mRNAs, of which three to four thousand mRNAs are translationally repressed. The timings and sites of translational activation of these dormant mRNAs are crucial for promoting oocyte maturation and embryonic development. How these mRNAs are accumulated and distributed in oocytes is therefore a fundamental issue to be explored. A method that enables visualization of mRNA molecules with high resolution in a simple manner would be valuable for understanding how oocytes accumulate and regulate the dormant mRNAs. We have developed a highly sensitive whole-mount in situ hybridization method using in vitro-synthesized RNA probes and the tyramide signal amplification (TSA) system optimized for mouse oocytes and embryos. By using this method, Pou5f1/Oct4, Emi2, and cyclin B1 mRNAs were detected in immature oocytes and 2-cell stage embryos. Confocal microscopy showed that these mRNAs formed granular structures in the oocyte cytoplasm. The structures of Pou5f1/Oct4 and cyclin B1 mRNAs persisted in 2-cell stage embryos. Pou5f1/Oct4 RNA granules exhibited a solid-like property in immature oocytes and became liquid-like droplets in 2-cell stage embryos. Double-staining of cyclin B1 mRNA with Emi2 or Pou5f1/Oct4 mRNA revealed that these mRNAs were distributed as different RNA granules without overlapping each other and that the size of cyclin B1 RNA granules tended to be larger than that of Emi2 RNA granules. The structures and distribution patterns of these mRNAs were further analyzed by N-SIM super-resolution microscopy. This analysis revealed that the large-sized RNA granules consist of many small-sized granules, suggesting the accumulation and regulation of dormant mRNAs as basal-sized RNA granules. The method established in this study can easily visualize the structure and distribution of mRNAs accumulated in mammalian oocytes and embryos with high sensitivity and super-resolution. This method is useful for investigating the cellular and molecular mechanisms of translational control of mRNAs by which maturation and early developmental processes are promoted.
Keywords: Embryo; Mammal; Maternal mRNA; Oocyte; Super-resolution microscopy; in situ hybridization.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Formation of mos RNA granules in the zebrafish oocyte that differ from cyclin B1 RNA granules in distribution, density and regulation.Eur J Cell Biol. 2016 Dec;95(12):563-573. doi: 10.1016/j.ejcb.2016.10.001. Epub 2016 Oct 4. Eur J Cell Biol. 2016. PMID: 27756483
-
Posttranscriptional regulation of maternal Pou5f1/Oct4 during mouse oogenesis and early embryogenesis.Histochem Cell Biol. 2020 Dec;154(6):609-620. doi: 10.1007/s00418-020-01915-4. Epub 2020 Sep 15. Histochem Cell Biol. 2020. PMID: 32930837
-
Pumilio1 phosphorylation precedes translational activation of its target mRNA in zebrafish oocytes.Zygote. 2018 Oct;26(5):372-380. doi: 10.1017/S0967199418000369. Epub 2018 Oct 5. Zygote. 2018. PMID: 30289101
-
Regulation of Translationally Repressed mRNAs in Zebrafish and Mouse Oocytes.Results Probl Cell Differ. 2017;63:297-324. doi: 10.1007/978-3-319-60855-6_13. Results Probl Cell Differ. 2017. PMID: 28779323 Review.
-
Sculpting the Transcriptome During the Oocyte-to-Embryo Transition in Mouse.Curr Top Dev Biol. 2015;113:305-49. doi: 10.1016/bs.ctdb.2015.06.004. Epub 2015 Jul 29. Curr Top Dev Biol. 2015. PMID: 26358877 Review.
Cited by
-
Spatial and quantitative gene expression analysis of SREB receptors in the gonads of green-spotted pufferfish (Dichotomyctere nigroviridis).Gen Comp Endocrinol. 2025 Jan 1;360:114641. doi: 10.1016/j.ygcen.2024.114641. Epub 2024 Nov 12. Gen Comp Endocrinol. 2025. PMID: 39536984
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

